Copyright
©2010 Baishideng Publishing Group Co.
World J Biol Chem. May 26, 2010; 1(5): 151-159
Published online May 26, 2010. doi: 10.4331/wjbc.v1.i5.151
Published online May 26, 2010. doi: 10.4331/wjbc.v1.i5.151
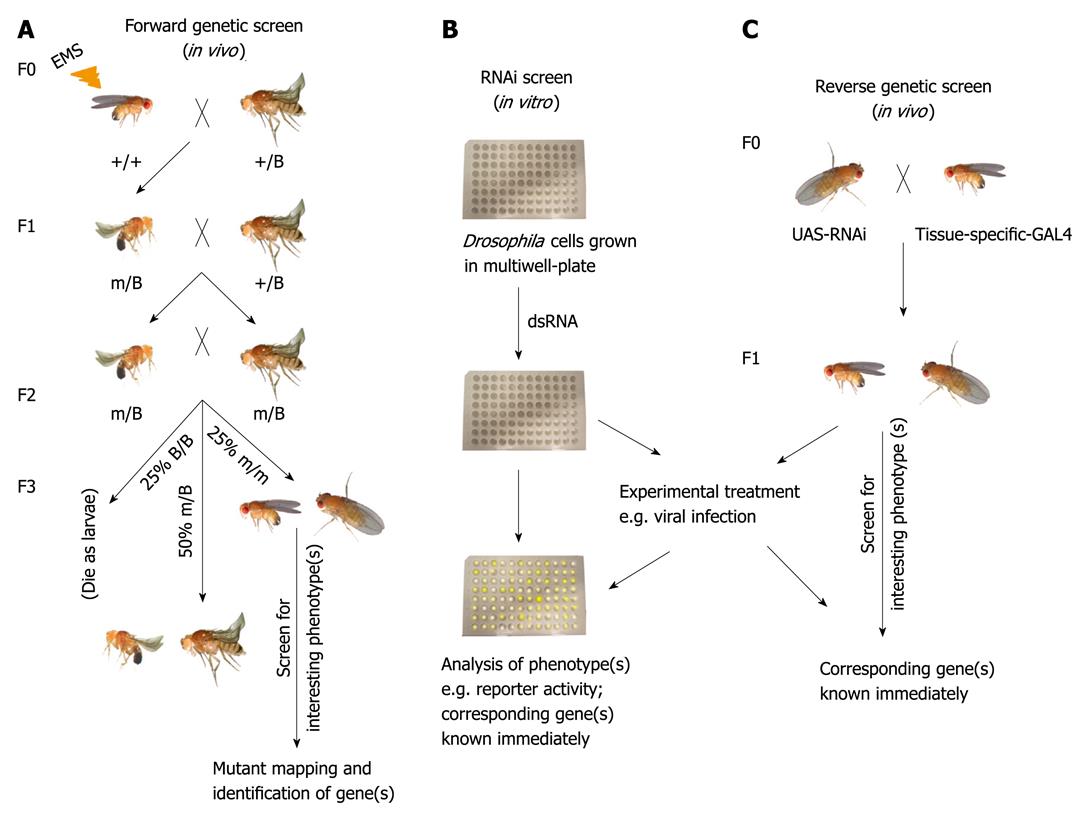
Figure 1 Methods for the systematic study of Drosophila gene function.
A: Forward genetic screen. Male flies subjected to ethyl methane sulphonate (EMS)-induced mutagenesis are crossed to virgin females that carry a balancer for the chromosome (indicated as B) to be screened. In the F1 progeny, each male inherits a mutagenized chromosome with a different spectrum of mutations. Individual F1 males are backcrossed to balancer stock. F2 male and F2 female carrying the same mutagenized chromosome are crossed with each other. 25% of the F3 are homozygous for the mutagenized chromosome. These are screened for interesting phenotype(s) followed by mapping and identification of corresponding genes; B: In vitro RNA interference (RNAi) screen. Drosophila cells are grown in multiwell-plates and incubated with known double-stranded RNA (dsRNA) to knock-down a specific gene. Thereafter, RNAi-treated cells are subjected to selected experimental procedures (e.g. viral infection). Interesting phenotype(s) can be immediately linked to the corresponding gene; C: Reverse genetic screen. Flies carrying dsRNA insertion for a particular gene driven by UAS promoter is crossed to a tissue-specific driver fly line. In the F1 progeny, the expressed GAL4 bind to UAS to drive the expression of the dsRNA to silence the expression of the selected gene. Flies are then tested in selected experimental conditions (for example viral infection).
-
Citation: Wang JH, Valanne S, Rämet M.
Drosophila as a model for antiviral immunity. World J Biol Chem 2010; 1(5): 151-159 - URL: https://www.wjgnet.com/1949-8454/full/v1/i5/151.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v1.i5.151