Copyright
©The Author(s) 2025.
World J Diabetes. Apr 15, 2025; 16(4): 97201
Published online Apr 15, 2025. doi: 10.4239/wjd.v16.i4.97201
Published online Apr 15, 2025. doi: 10.4239/wjd.v16.i4.97201
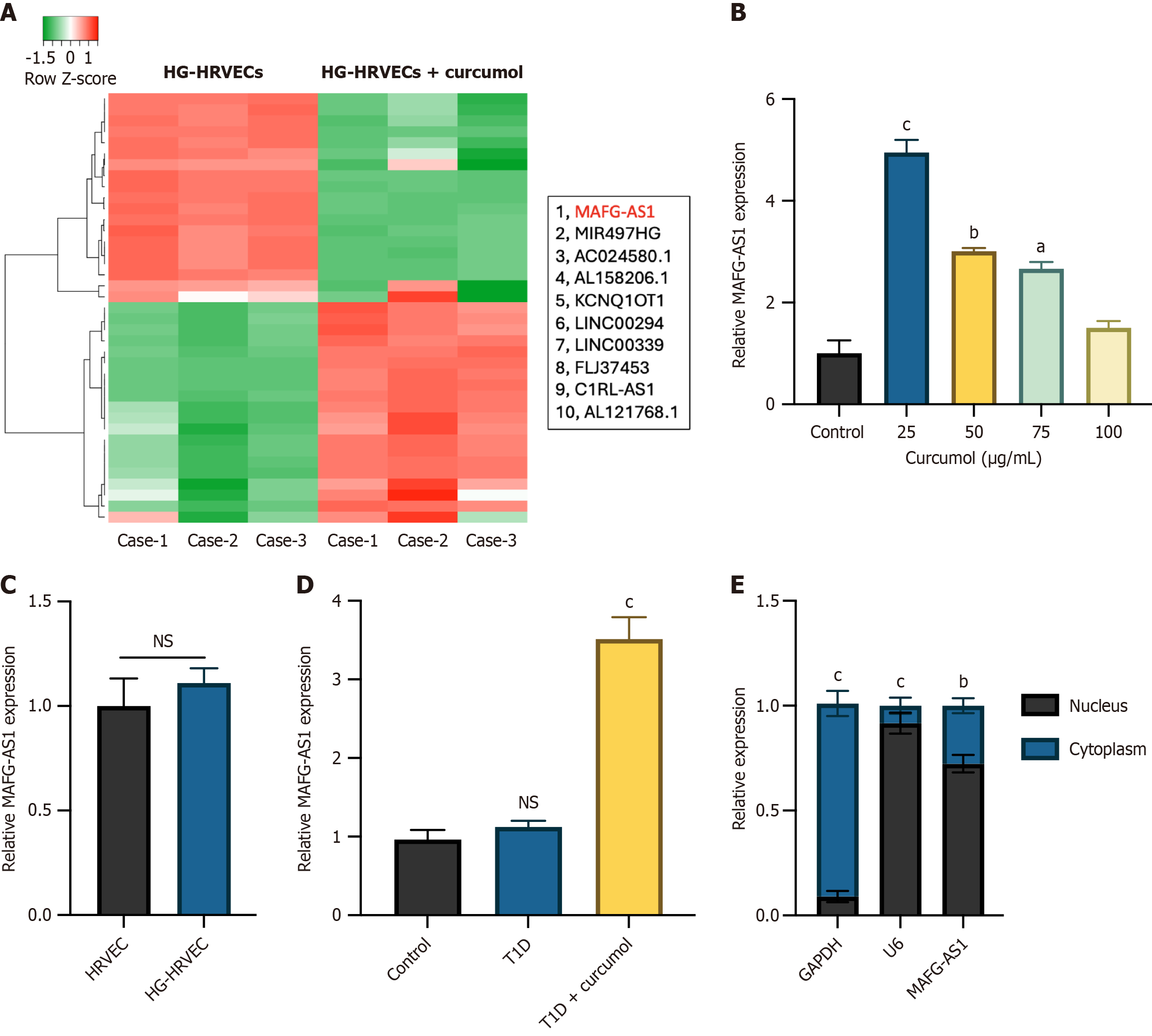
Figure 3 Identification of dysregulated long non-coding RNAs upon curcumol treatment.
A: Long non-coding RNA (lncRNA) microarray analysis was utilized to identify significantly dysregulated lncRNAs expression in high glucose-human retinal vascular endothelial cells (HG-HRVECs) and curcumol-treated HG-HRVECs. The top 10 upregulated lncRNAs are shown; B: LncRNA MAF transcription factor G antisense RNA 1 (MAFG-AS1) expression in HG-HRVECs and HG-HRVECs treated with different doses of curcumol was measured by quantitative real-time polymerase chain reaction (qRT-PCR); C: MAFG-AS1 expression in HRVECs and HG-HRVECs was detected by qRT-PCR; D: MAFG-AS1 expression the retinal tissues of control mice, type 1 diabetes mice, and type 1 diabetes mice treated with curcumol was detected by qRT-PCR; E: Cellular distribution of MAFG-AS1 in HRVECs. Data represent the mean ± SD. aP < 0.05, bP < 0.01, cP < 0.001, NS: No significance. HG-HRVECs: High glucose-human retinal vascular endothelial cells; MAFG-AS1: MAF transcription factor G antisense RNA 1; T1D: Type 1 diabetes; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
- Citation: Rong H, Hu Y, Wei W. Curcumol ameliorates diabetic retinopathy via modulating fat mass and obesity-associated protein-demethylated MAF transcription factor G antisense RNA 1. World J Diabetes 2025; 16(4): 97201
- URL: https://www.wjgnet.com/1948-9358/full/v16/i4/97201.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i4.97201