Copyright
©The Author(s) 2025.
World J Diabetes. Apr 15, 2025; 16(4): 100113
Published online Apr 15, 2025. doi: 10.4239/wjd.v16.i4.100113
Published online Apr 15, 2025. doi: 10.4239/wjd.v16.i4.100113
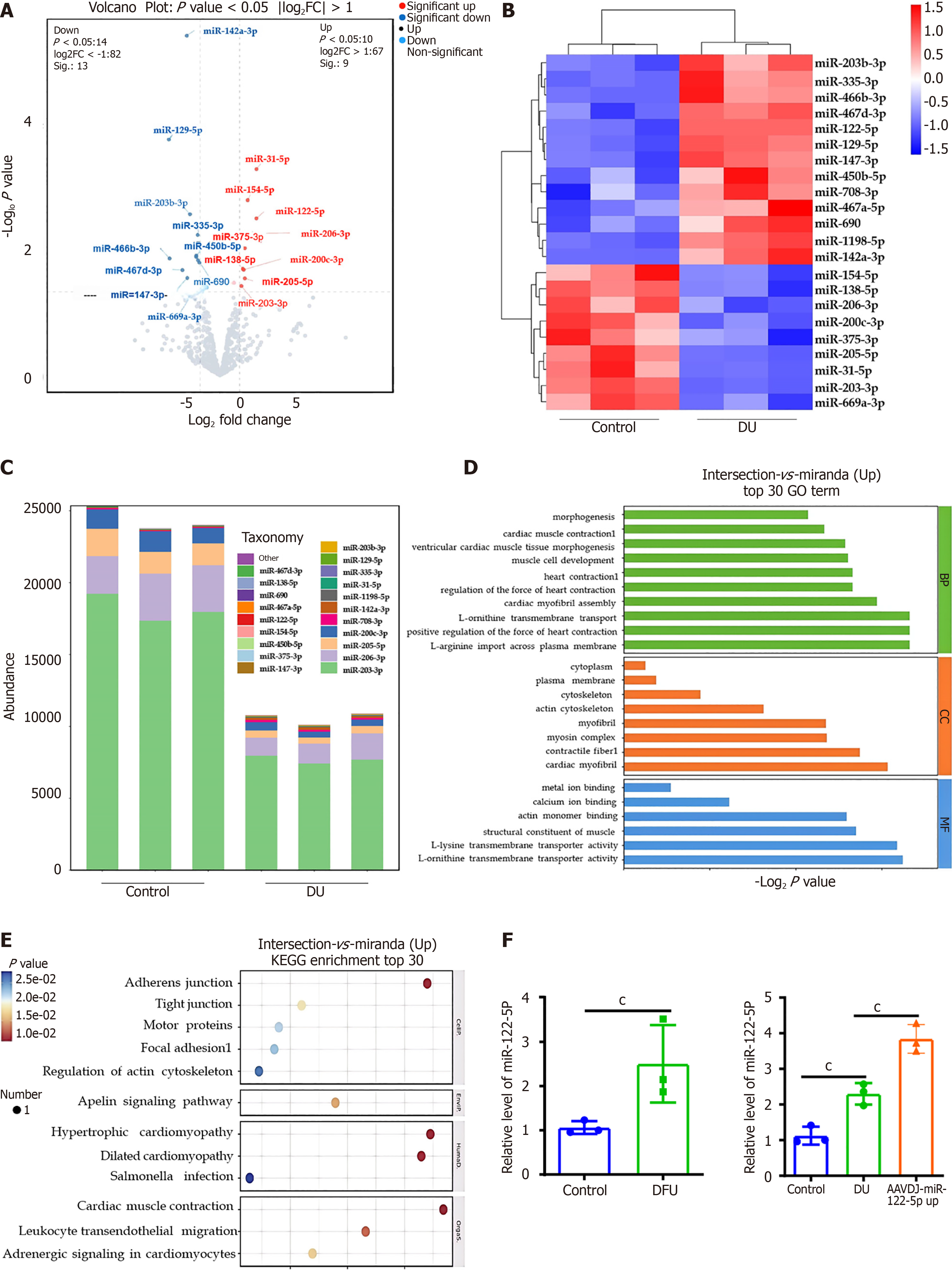
Figure 1 Sequencing and bioinformatics analysis of MicroRNAs.
A: In diabetic ulcer mice wounds, wound tissues were harvested 14 days postinjury, and total RNA was extracted. Subsequently, MicroRNAs (miR)-seq was performed, followed by the generation of a volcano plot; B: Representative heatmap of differentially expressed miRs in the skin tissues of diabetic ulcer mice; C: The top 10 miRs with the highest expression levels in each group; D: Gene ontology annotations and enrichment analysis of the target genes of miRs; E: Kyoto encyclopedia of genes and genomes annotations and enrichment analysis of the target genes of miRs; F: Relative expression levels of miR-122-5p in wound tissue from streptozotocin-induced diabetic mice [2-∆∆Ct = 2.30, 95% confidence interval (CI): 1.81-2.72] showed a significant increase compared to normal mice (2-∆∆Ct = 1.12, 95%CI: 0.64-1.31, P < 0.001) by quantitative real-time polymerase chain reaction; Further experiments demonstrated that the miR-122-5p overexpression group (2-∆∆Ct = 3.82, 95%CI: 3.45-4.37, P < 0.001) exhibited significantly higher expression compared to the diabetic ulcer group; miR-122-5p levels were significantly increased in patients with diabetic foot ulcer (2-∆∆Ct = 1.86, 95%CI: 2.14-3.51) compared with healthy individuals (2-∆∆Ct = 1, 95%CI: 0.96-1.23, P < 0.001). cP < 0.001. miR: MicroRNA; FC: Fold change; DU: Diabetic ulcer; GO: Gene ontology; KEGG: Kyoto encyclopedia of genes and genomes; DFU: Diabetic foot ulcer; AAVDJ: Adeno-associated virus-DJ.
- Citation: Yuan MJ, Huang HC, Shi HS, Hu XM, Zhao Z, Chen YQ, Fan WJ, Sun J, Liu GB. MicroRNA-122-5p is upregulated in diabetic foot ulcers and decelerates the transition from the inflammatory to the proliferative stage. World J Diabetes 2025; 16(4): 100113
- URL: https://www.wjgnet.com/1948-9358/full/v16/i4/100113.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i4.100113