Copyright
©The Author(s) 2025.
World J Diabetes. Feb 15, 2025; 16(2): 101538
Published online Feb 15, 2025. doi: 10.4239/wjd.v16.i2.101538
Published online Feb 15, 2025. doi: 10.4239/wjd.v16.i2.101538
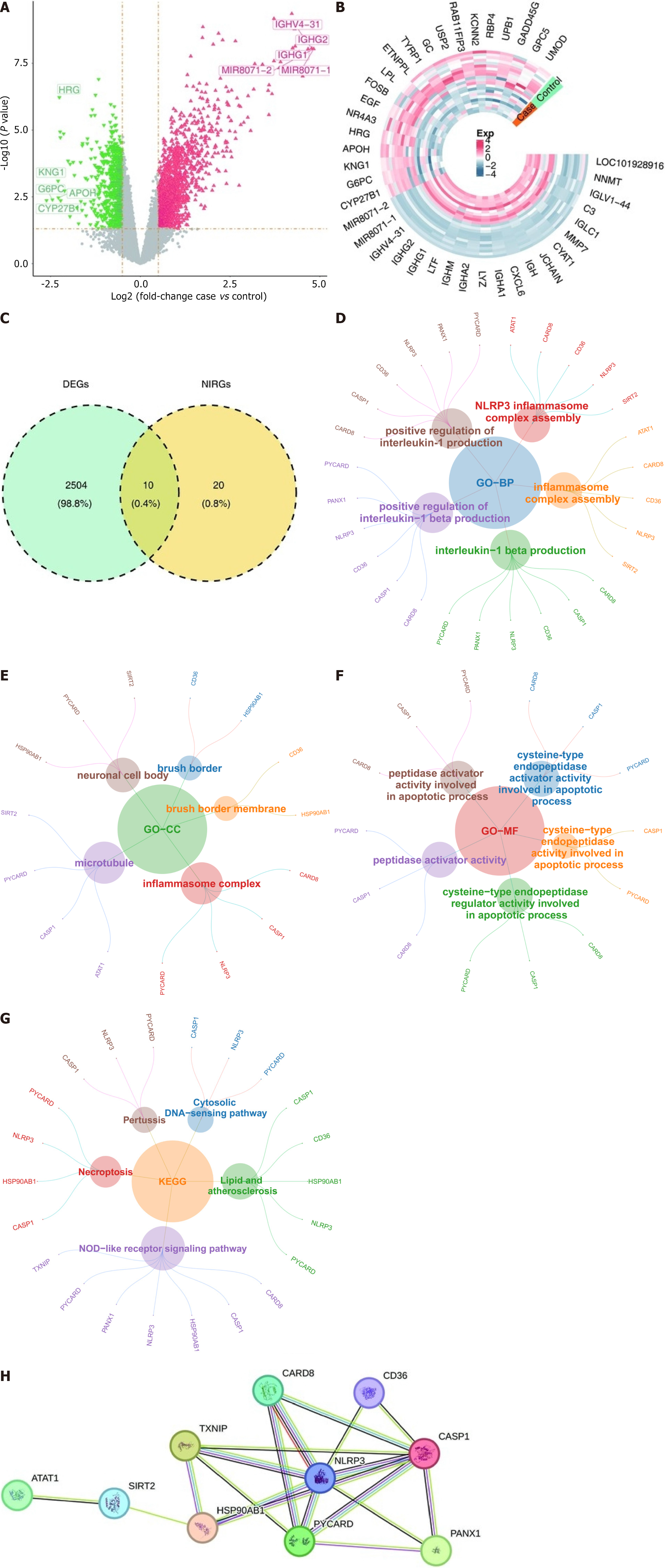
Figure 1 The 10 intersection genes were enriched in the NOD-like receptor signaling pathway.
A: Volcano plot of differentially expressed genes (DEGs); B: Heatmap of DEGs; C: Venn diagram of the intersection of DEGs and NLRP3 inflammasome-associated genes; D-F: Gene Ontology analysis of the intersection genes, including molecular function, cellular components, and biological processes; G: Significantly enriched Kyoto Encyclopedia of Genes and Genomes pathway analysis of the intersecting genes; H: Protein-protein interaction network of DEGs. DEGs: Differentially expressed genes; NLRP3: NOD-like receptor thermal protein domain associated protein 3; NIRGs: NLRP3 inflammasome-associated genes; GO: Gene Ontology; BP: Biological process; CC: Cellular component; MF: Molecular function; KEGG: Kyoto Encyclopedia of Genes and Genomes; SIRT2: Sirtuin 2; CASP1: Caspase 1.
- Citation: Zhou Y, Fang X, Huang LJ, Wu PW. Transcriptome and single-cell profiling of the mechanism of diabetic kidney disease. World J Diabetes 2025; 16(2): 101538
- URL: https://www.wjgnet.com/1948-9358/full/v16/i2/101538.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i2.101538