Copyright
©The Author(s) 2023.
World J Diabetes. Jul 15, 2023; 14(7): 1077-1090
Published online Jul 15, 2023. doi: 10.4239/wjd.v14.i7.1077
Published online Jul 15, 2023. doi: 10.4239/wjd.v14.i7.1077
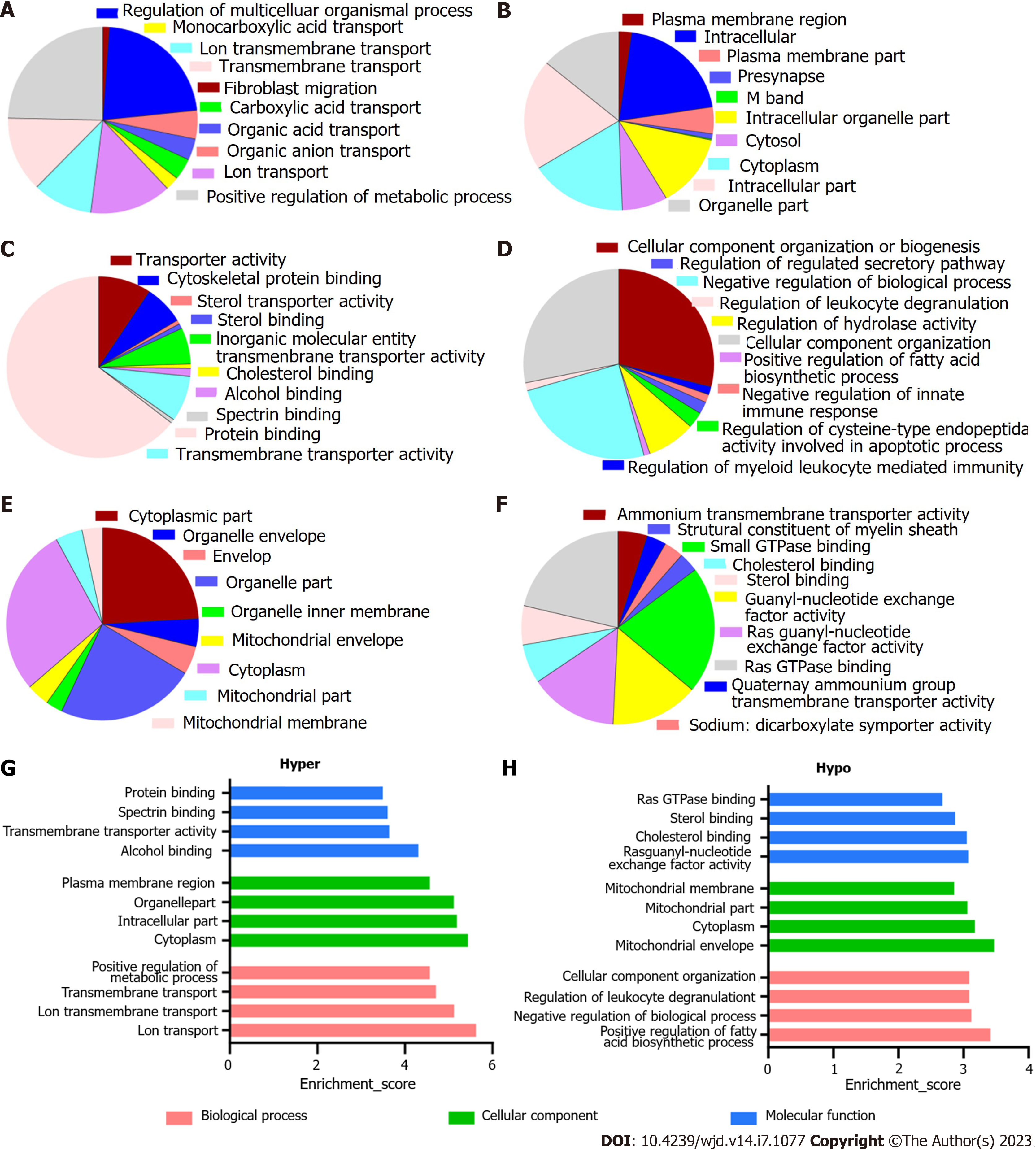
Figure 3 Overall distribution of Gene Ontology analysis.
A-C: Classification of hypermethylated mRNAs in the biological process (BP), cellular component (CC), and molecular function (MF) categories. Among the enriched Gene Ontology (GO) terms, 580 BPs, 110 CCs, and 100 MFs had higher mRNA methylation levels; D-F: Classification of hypomethylated mRNAs in the BP, CC, and MF categories. For the hypomethylated mRNAs, 288 BPs, 47 CCs, and 67 MFs were identified; G: The top four most enriched GO terms of the hypermethylated mRNAs; H: The top four most enriched GO terms of the hypomethylated mRNAs.
- Citation: Cai L, Han XY, Li D, Ma DM, Shi YM, Lu Y, Yang J. Analysis of N6-methyladenosine-modified mRNAs in diabetic cataract. World J Diabetes 2023; 14(7): 1077-1090
- URL: https://www.wjgnet.com/1948-9358/full/v14/i7/1077.htm
- DOI: https://dx.doi.org/10.4239/wjd.v14.i7.1077