Copyright
©The Author(s) 2025.
World J Gastrointest Oncol. Apr 15, 2025; 17(4): 103048
Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103048
Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103048
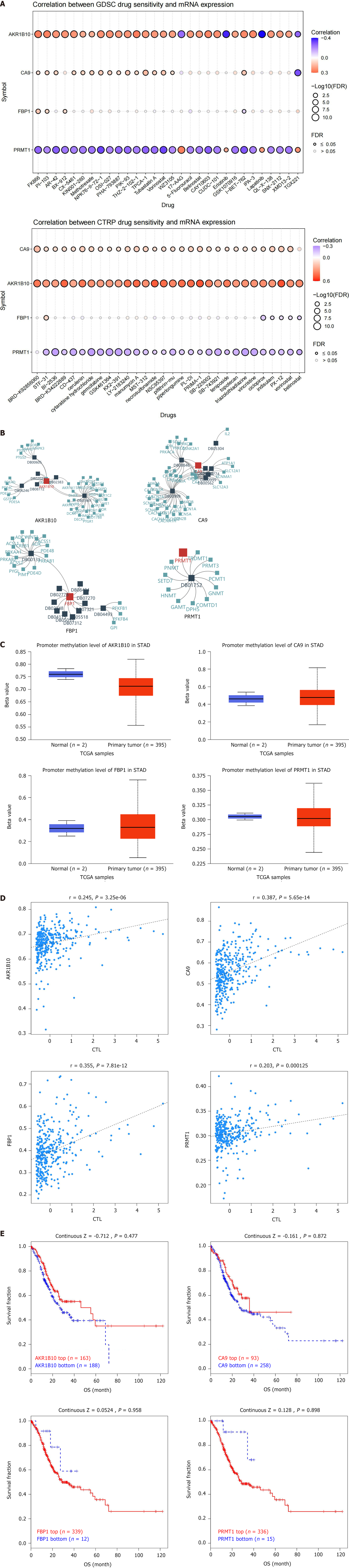
Figure 9 Hub genes are involved in epigenetic regulation and repair of damaged genes.
A: Correlation of hub genes with immunotherapy; B: Network diagram of hub genes and immunotherapy; C: Expression level map of hub gene methylation. Blue and red represent the normal and tumor groups, respectively; D: Correlation between hub gene methylation levels and cytotoxic T lymphocyte markers; E: Survival curves of hypermethylated and hypomethylated subgroups of hub genes. CTRP: Cancer Therapeutics Response Portal; CA9: Carbonic anhydrase 9; FBP1: Fructose-bisphosphatase 1; PRMT1: Protein arginine methyltransferase 1; AKR1B10: Aldo-keto reductase family 1 member B10; TCGA: The Cancer Genome Atlas; CTL: Cytotoxic T lymphocyte; OS: Overall survival.
- Citation: Meng FD, Jia SM, Ma YB, Du YH, Liu WJ, Yang Y, Yuan L, Nan Y. Identification of key hub genes associated with anti-gastric cancer effects of lotus plumule based on machine learning algorithms. World J Gastrointest Oncol 2025; 17(4): 103048
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/103048.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.103048