Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Jul 15, 2024; 16(7): 3032-3054
Published online Jul 15, 2024. doi: 10.4251/wjgo.v16.i7.3032
Published online Jul 15, 2024. doi: 10.4251/wjgo.v16.i7.3032
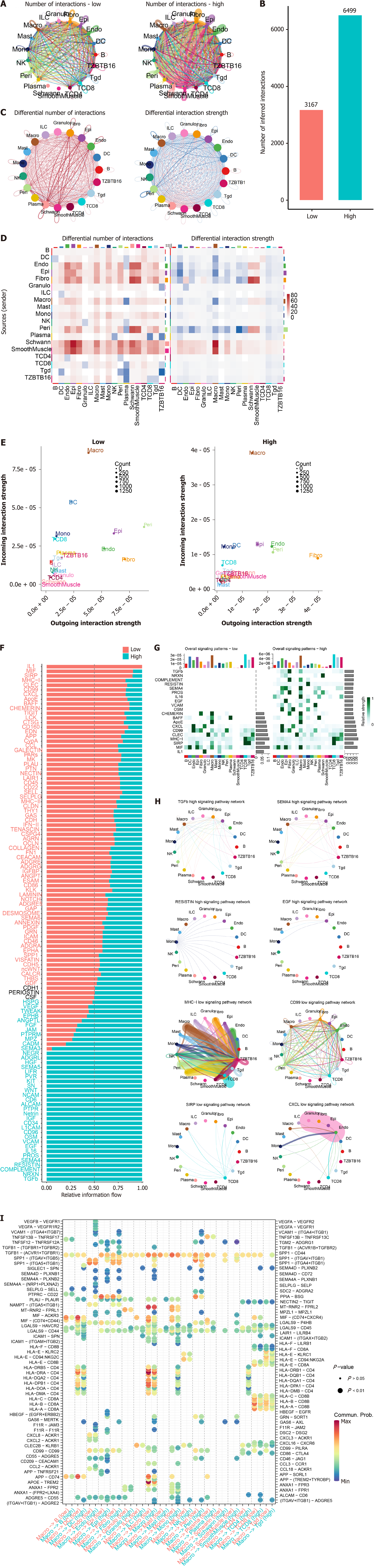
Figure 8 Cell-cell interaction and the signature.
A: Although most interactions exist in both low and high-risk samples; B: The interaction number is significantly different; C and D: Despite that the interaction number is higher in high-risk samples, the interaction strength is lower; E: The macrophage is noticed as the most significantly altered immune cell among the interactions; F: The pathway level interaction information flow was visualized for both groups, overall; G: Top 10 for each group; H: Among these interactions, it is noticed that macrophage is especially active in high-risk samples while interactions involved CD8+ T cell were increased in the low-risk group; I: Specific pathways and cell type interaction landscape also revealed that macrophage is activated in high-risk samples.
- Citation: Liu HX, Feng J, Jiang JJ, Shen WJ, Zheng Y, Liu G, Gao XY. Integrated single-cell and bulk RNA sequencing revealed an epigenetic signature predicts prognosis and tumor microenvironment colorectal cancer heterogeneity. World J Gastrointest Oncol 2024; 16(7): 3032-3054
- URL: https://www.wjgnet.com/1948-5204/full/v16/i7/3032.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i7.3032