Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Jun 15, 2024; 16(6): 2571-2591
Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2571
Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2571
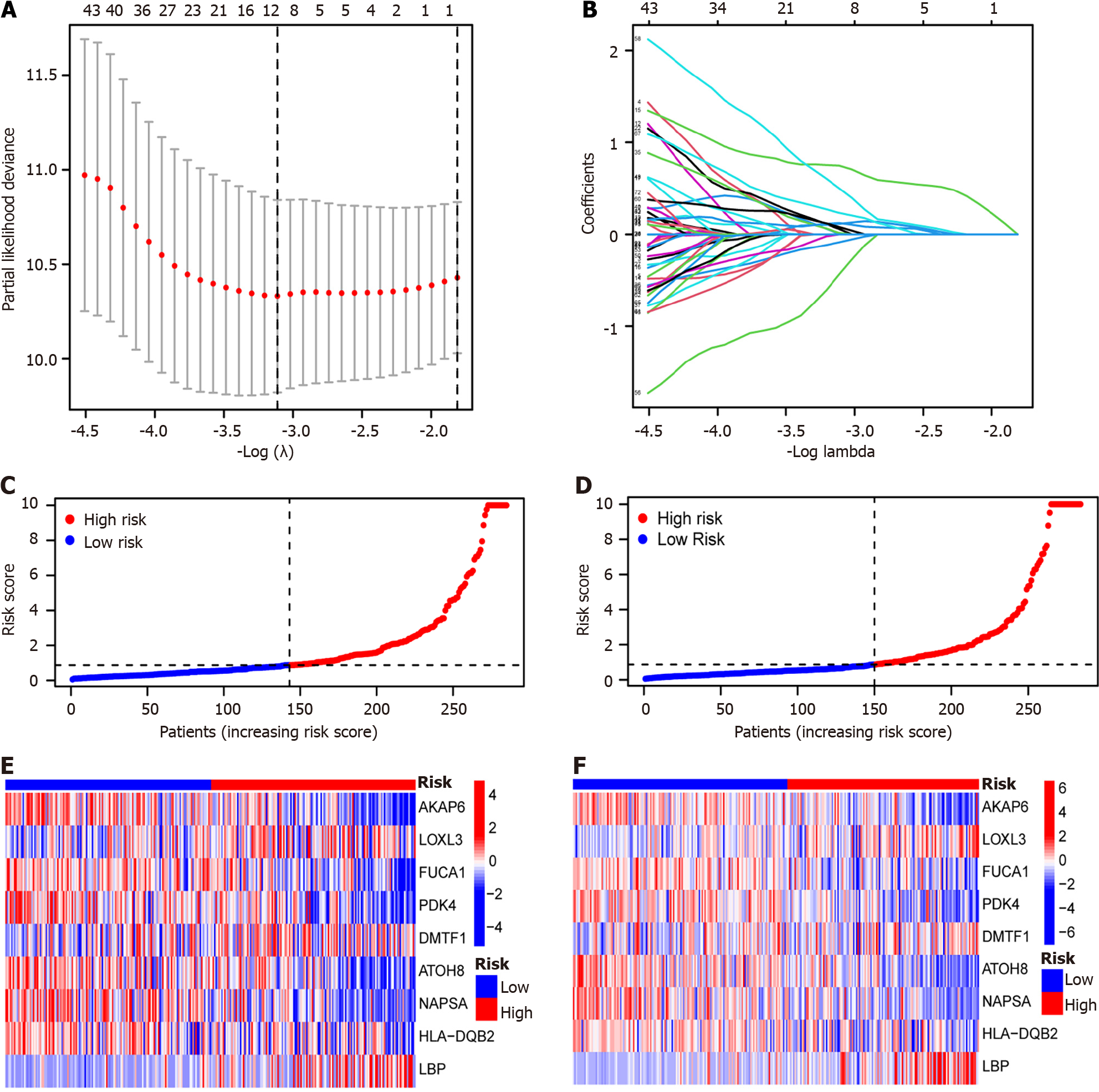
Figure 2 Risk grouping construction based on The Cancer Genome Atlas-Liver Hepatocellular Carcinoma, GSE25097, and GSE36376 combined datasets.
A: Least absolute shrinkage and selection operator is a gradient descent method for determining hub genes; B: Number of hub genes and their corresponding coefficients; C: Train group samples were classified into high- and low-risk groups based on the risk scores. Red indicates high-risk group samples, and blue indicates low-risk group samples; D: Test group samples have been classified into high-risk and low-risk groups based on the risk scores. Red indicates high-risk group samples, and blue indicates low-risk group samples; E: The hub genes expression within the high- and low-risk groups of the train group. Red indicates up-regulated expression, blue indicates under-expression of the expression, and white indicates zero expression; F: The hub genes expression within high- and low-risk groups of the test group. Red indicates up-regulated expression, blue indicates under-expression of the expression, and white indicates zero expression.
- Citation: Ren QS, Sun Q, Cheng SQ, Du LM, Guo PX. Hepatocellular carcinoma: An analysis of the expression status of stress granules and their prognostic value. World J Gastrointest Oncol 2024; 16(6): 2571-2591
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2571.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2571