Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. May 15, 2024; 16(5): 2060-2073
Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2060
Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2060
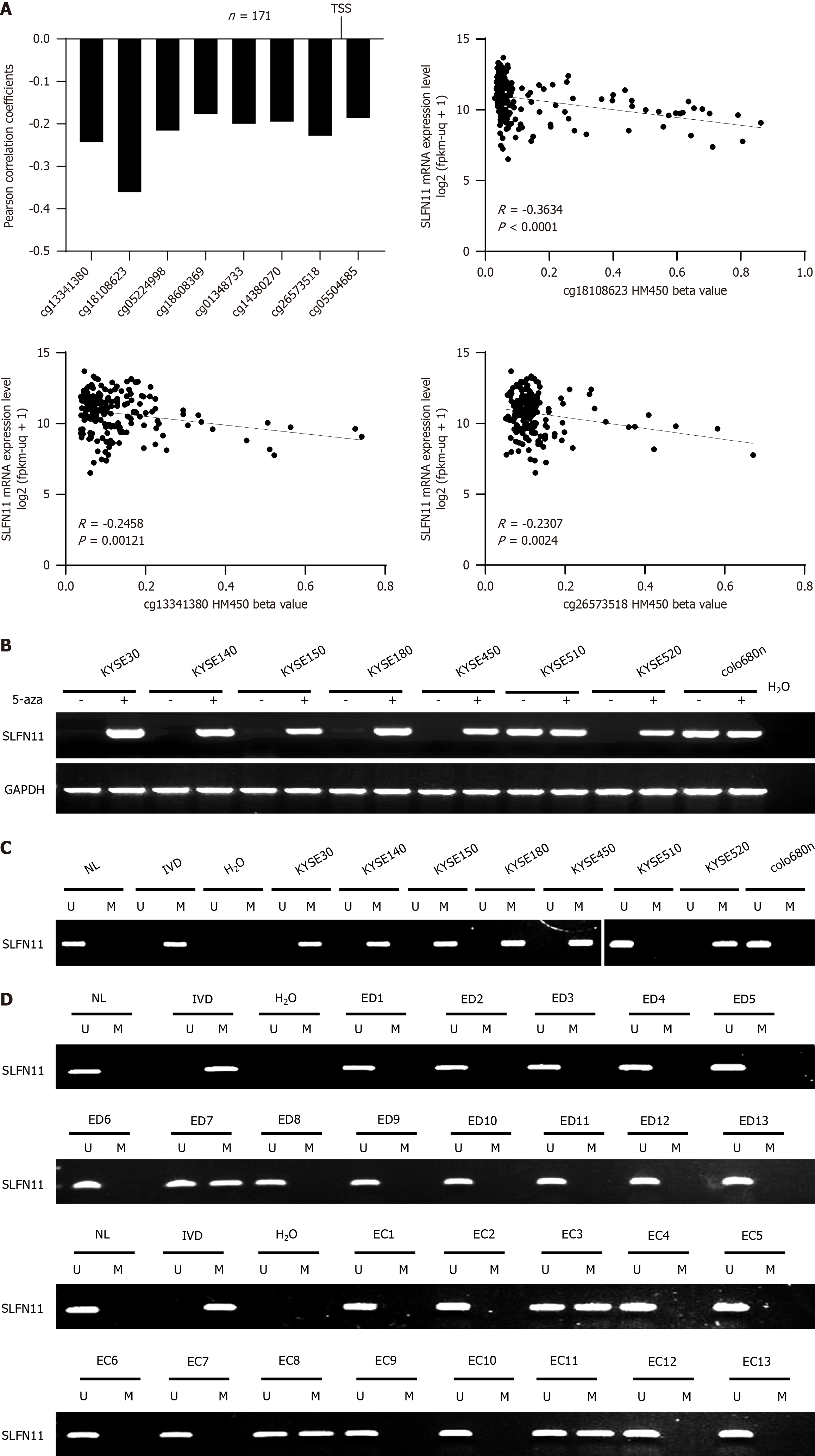
Figure 1 The expression and methylation status of Schlafen-11 in esophageal squamous cell carcinoma.
A: Correlation analysis between Schlafen-11 (SLFN11) mRNA expression and methylation levels of 8 CpG sites around transcription start site retrieved from TCGA datasets (n = 171). Scatter plots shown inverse relevance of SLFN11 expression and methylation status in representative CpG sites (cg18108623, cg13341380, and cg26573518); B: Semi-quantitative RT-PCR showing the expression of SLFN11 in esophageal squamous cell carcinoma (ESCC) cell lines before and after treatment with 5-aza-2’-deoxycytidine (5-aza); C: Detection of the methylation status of SLFN11 by methylation-specific polymerase chain reaction (MSP) in ESCC cells; D: Representative MSP results of SLFN11 in esophageal tissue samples. TSS: Transcription start site; KYSE30, KYSE140, KYSE150, KYSE180, KYSE450, KYSE510, KYSE520, and colo680n are ESCC cells; 5-aza: 5-aza-2’-deoxycytidine; GAPDH: Internal control of RT-PCR; H2O: Double distilled water; (-): Absence of 5-aza; (+): Administration of 5-aza; U: Unmethylated alleles; M: Methylated alleles; IVD: In vitro methylated DNA as methylation control; NL: Normal peripheral lymphocytes DNA as unmethylation control; ED: Esophageal dysplasia; EC: Esophageal squamous cell carcinoma.
- Citation: Zhou J, Zhang MY, Gao AA, Zhu C, He T, Herman JG, Guo MZ. Epigenetic silencing schlafen-11 sensitizes esophageal cancer to ATM inhibitor. World J Gastrointest Oncol 2024; 16(5): 2060-2073
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/2060.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.2060