Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. May 15, 2024; 16(5): 2018-2037
Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2018
Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2018
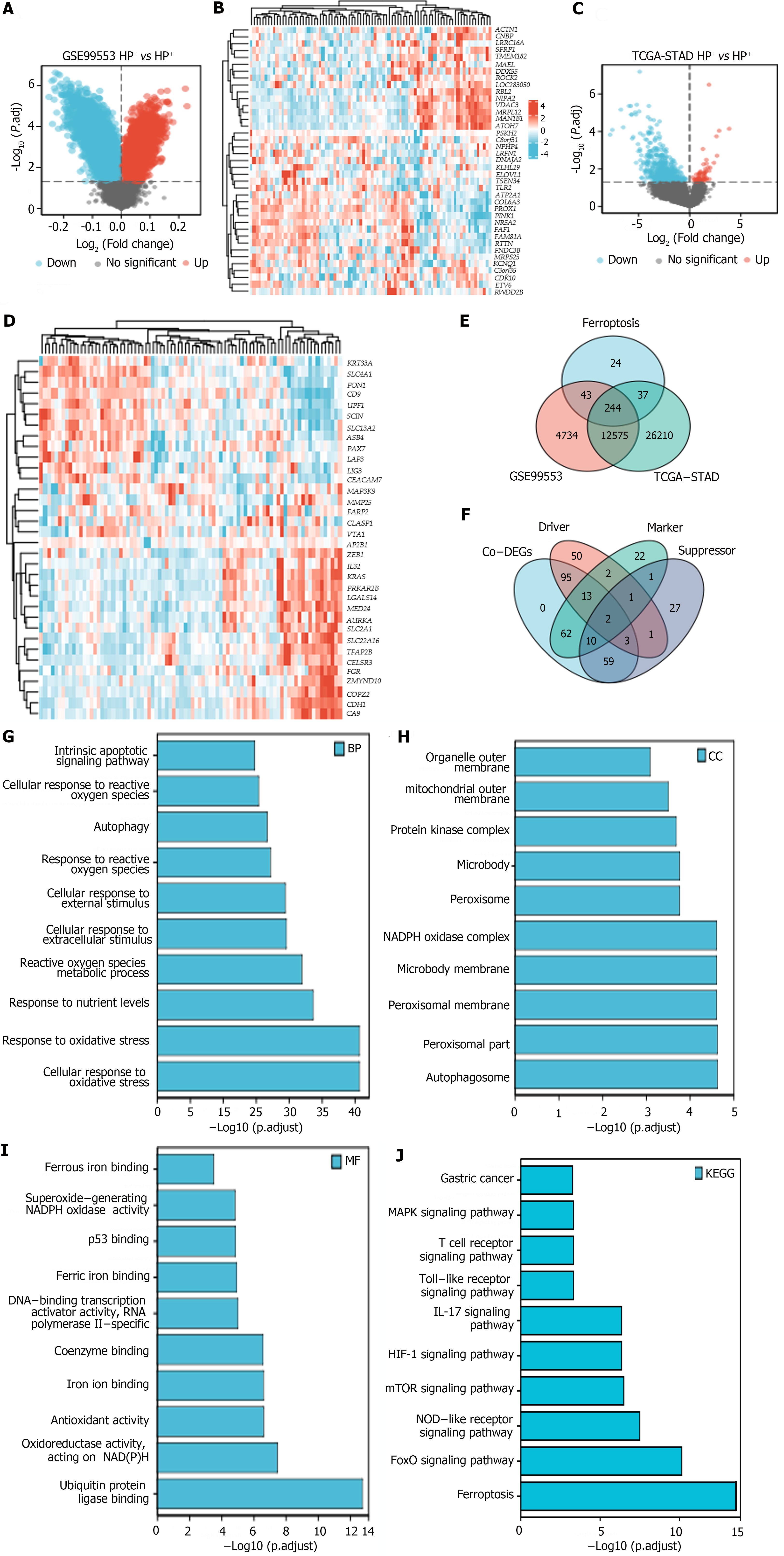
Figure 2 Identification of co-regulated differentially expressed genes and GO and KEGG analyses.
A: Differential gene expression map in GSE99553; B: Heatmap of differentially expressed genes in GSE99553; C: Differential gene expression map in the cancer genome atlas of stomach adenocarcinoma (TCGA-STAD) cohort; D: Heatmap of differentially expressed genes in the TCGA-STAD cohort; E and F: Overlapping differentially expressed genes in TCGA-STAD and GSE99553 related to ferroptosis; G-J: GO and KEGG analysis results of the Co-DEGs. NADPH: Nicotinamide adenine dinucleotide phosphate; MAPK: Mitogen-activated protein kinase; HIF-1: Hypoxia-inducible factor; IL-17: Interleukin-17; FoxO: Forkhead box O; NOD: Nucleotide-binding oligomerization domain; TCGA-STAD: The cancer genome atlas of stomach adenocarcinoma; Co-DEGs: Co-regulated differentially expressed genes.
- Citation: Wang L, Gong WH. Predictive model using four ferroptosis-related genes accurately predicts gastric cancer prognosis. World J Gastrointest Oncol 2024; 16(5): 2018-2037
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/2018.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.2018