Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1104
Peer-review started: December 15, 2023
First decision: January 15, 2024
Revised: January 28, 2024
Accepted: February 26, 2024
Article in press: February 26, 2024
Published online: April 15, 2024
Esophageal cancer (EC) is the seventh most common cancer worldwide, and esophageal squamous cell carcinoma (ESCC) accounts for the majority of cases of EC. To effectively diagnose and treat ESCC and improve patient prognosis, timely diagnosis in the initial phase of the illness is necessary. This article offers a detailed summary of the latest advancements and emerging technologies in the timely identification of ECs. Molecular biology and epigenetics approaches involve the use of molecular mechanisms combined with fluorescence quanti
Core Tip: The incidence of esophageal cancer is high worldwide, with and esophageal squamous cell carcinoma (ESCC) is the predominant histological subtype. With in-depth research and the development of technology, various methods have been employed to promptly identify early-stage ESCC. This review will help to elucidate new advances and applications of diagnostic methods for early ESCC, including molecular biology, epigenetics, microbiology, endoscopy, and artificial intelligence approaches.
- Citation: Qi JH, Huang SL, Jin SZ. Novel milestones for early esophageal carcinoma: From bench to bed. World J Gastrointest Oncol 2024; 16(4): 1104-1118
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1104.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1104
Esophageal cancer (EC) was identified as the seventh most common type of carcinomatosis worldwide and accounted for the sixth highest number of cancer-related deaths in 2020. The classification of EC at the histological level primarily comprises two categories: Adenocarcinoma of the esophagus (EAC) and esophageal squamous cell carcinoma (ESCC)[1]. ESCC is widely prevalent worldwide, particularly in East Asia and certain regions of Africa[2,3]. Despite exceeding advancements in the diagnostic modalities and treatment strategies available for ESCC, effectively managing this condition remains a formidable challenge due to the absence of specific early-stage symptoms, the substantial potential for metastasis and recurrence, and resistance to traditional therapeutic approaches. The likelihood of patients with advanced-stage ESCC surviving is merely 10%-20% over a span of 5 years[4]. The timely identification and diagnosis of ESCC are of paramount importance for increasing the likelihood of eligibility for curative surgical resection[5].
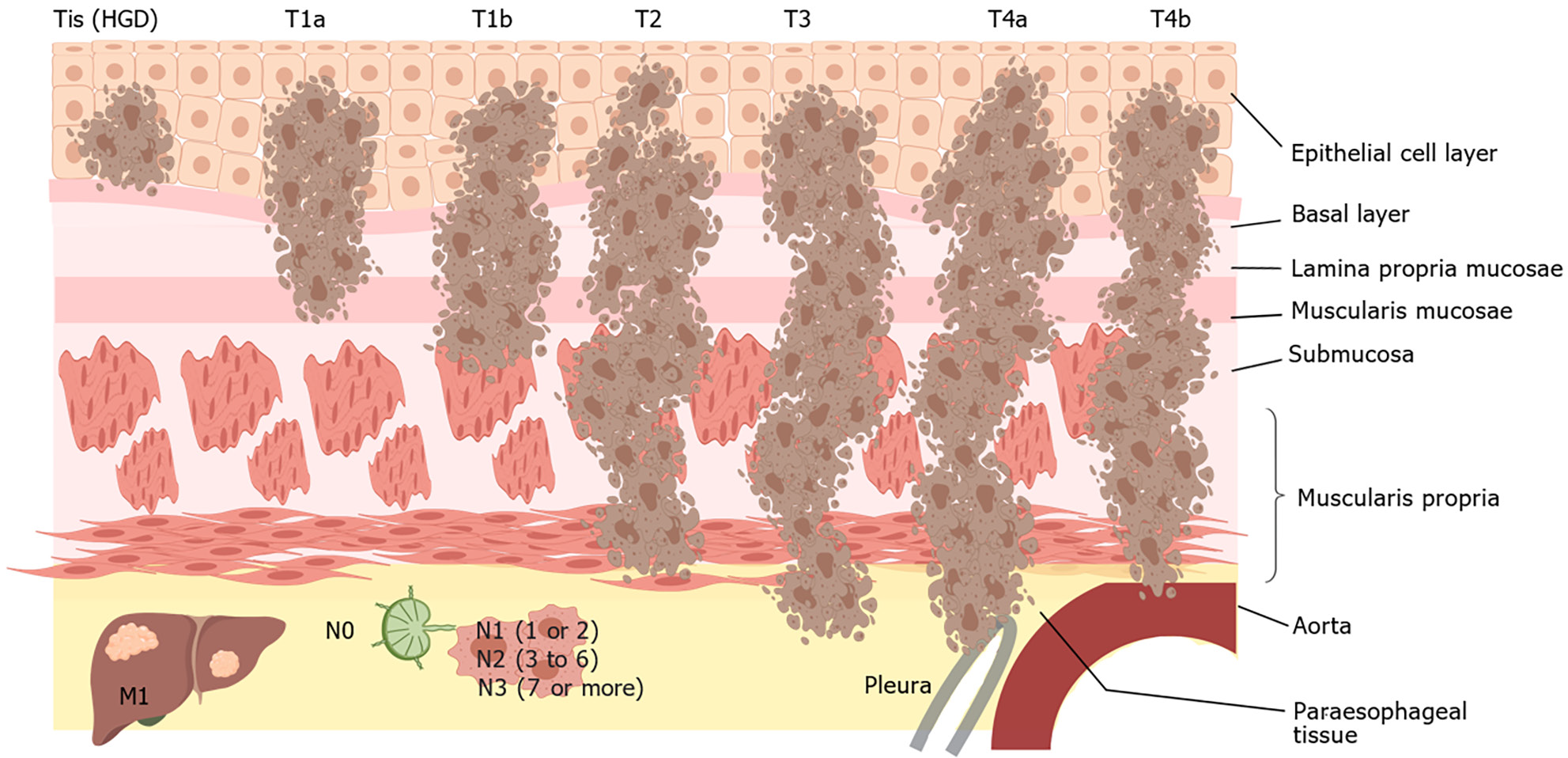
The etiology of ESCC is believed to be closely associated with factors such as tobacco use, consumption of alcoholic beverages, excessive intake of hot beverages, and inadequate nutrition. The potential for genetic and epigenetic changes within cells can be increased by prolonged exposure of the squamous epithelium in the upper gastrointestinal tract to different carcinogens, thus promoting tumorigenesis[6] (Figure 1). To explore the molecular abnormalities responsible for the progression of ESCC, a team of researchers has conducted comprehensive investigations via genomics, epigenomics, and transcriptomics to identify distinctive characteristics associated with this disease. The alterations frequently observed in tumors include amplification of genes that promote tumor growth and those that regulate the cell cycle and programmed cell death, and a high frequency of mutations is observed in tumors[7]. Together, mechanisms such as DNA methylation, histone modification, noncoding RNA regulation, chromatin remodeling, and nucleosome localization constitute epigenetic modifications. Studies have substantiated the impact of aberrant epigenetic processes on the transcription of genes associated with cellular growth, differentiation, programmed cell death, malignant transformation, and tumor progression. These findings have significant implications for clinical diagnosis, treatment strategies, and tumor prevention[8].
The human microbiota is a complex ecosystem of microorganisms. The nutrition, immunity, and metabolism of hosts are significantly influenced by the active participation of microbial communities, specifically bacteria, during coevolution[9]. Recently, the involvement of microorganisms has been found to be crucial in the pathogenesis and progression of gastrointestinal disorders. An accumulating body of evidence suggests that dysregulation of specific species contributes to the onset and progression of neoplasms through mechanisms such as the production of carcinogenic toxins, sup
Esophageal endoscopic biopsy combined with histological analysis is widely regarded as the most reliable method for early detection and diagnosis. Advanced endoscopic imaging techniques, such as chromoendoscopy, virtual chromoendoscopy, and magnifying endoscopy, can significantly enhance the sensitivity of detecting early-stage cancer[14]. Minimally invasive endoscope-guided treatment offers effective management for the majority of ESCCs and precancerous lesions, leading to a 90% chance of survival for up to 5 years[15]. However, due to the varying levels of expertise among endoscopists, there are still numerous cases in which the disease was not detected, resulting in missed diagnoses. Numerous studies have demonstrated the significant enhancement of diagnostic accuracy for lesions, particularly in the domain of computer-assisted detection for timely recognition of ESCC, due to the rapid advancement of artificial intelligence (AI)[16].
In summary, this review comprehensively examines recent advancements in the early diagnosis of ESCC. We prioritized the exploration and utilization of biomarkers, technological improvements in endoscopic examinations, and the potential role of AI in diagnosing ESCC. Hopefully, through these discussions, the enhancement of the precision and efficiency of early detection for this type of cancer can be facilitated by offering important perspectives.
The major histological subtypes of EC can be categorized into two groups. In Western developed countries, the prevalence of EAC surpasses that of ESCC, whereas in developing countries, the predominance of ESCC is greater, accounting for approximately 90% of all cancer cases[17,18]. ESCC cells mainly infiltrate the middle of the esophagus and have a high tendency for local and lymphatic spread[19]. EC diagnosis often occurs during the intermediate to advanced stages of disease, underscoring the crucial significance of timely detection and intervention for improving the prognosis of individuals afflicted with this disease.
Research has presented evidence of correlations between a wide range of environmental and genetic factors and risk factors for ESCC, including excessive alcohol consumption, hot food or beverages, a monotonous diet, nutritional deficiencies, micronutrient deficiencies, excessive consumption of red meat, processed meats, and pickled vegetables. Factors that have been recognized to reinforce the progression of malignant neoplasms include exposure to chemical carcinogens such as nitrosamines and polycyclic aromatic hydrocarbons, Human papillomavirus, a family history of cancer, and genetic alterations[20-22].
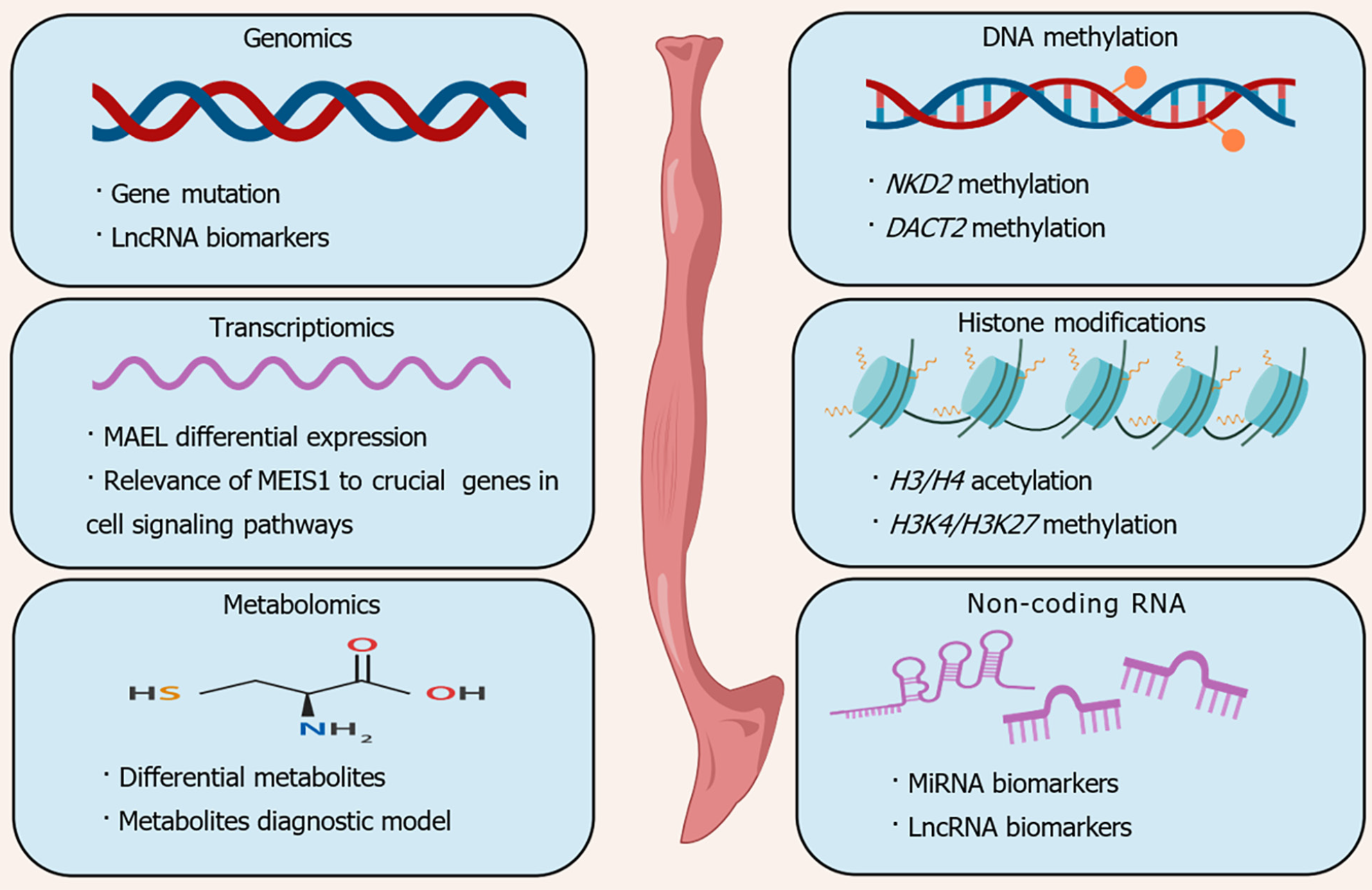
Molecular biology research techniques and laboratory diagnostic tools are designed to study targets or molecular markers of diseases through the acquisition, identification, and analysis of genetic material. In the field of cancer research, molecular biomarkers are substances detected within the body that serve as indicators of the presence of cancer. Typical indicators include variances in genetic makeup, disparities in mRNA and protein expression patterns, posttranslational alterations of proteins, and alterations in the levels of metabolites[23]. The alteration of molecules occurring during tumor progression over an extended period enables the utilization of fundamental molecular biology techniques for cancer diagnosis, prognosis determination, and disease progression monitoring (Figure 2).
Alterations in the genomic DNA sequence of cancer cells are a primary contributor to the development of carcinomas. With the emergence of novel technologies, it has become feasible to acquire a substantial quantity of comprehensive DNA sequences from cancer genomes for the purpose of detecting alterations in both normal and cancerous genomes. Furthermore, analysis of the genome revealed changes in the DNA sequences found within tumor DNA samples, including insertions and deletions, copy number variations, structural rearrangements, and a reduction in heterozygosity[24].
The widespread utilization of whole-exome sequencing (WES) as a prominent technical method has been facilitated by continuous progress in next-generation sequencing within the realm of human genomics. Urabe et al[25] used WES to analyze DNA from tumor tissue and paired healthy tissue from ESCC patients and identified mutated somatic genes. TP53 exhibited the highest mutation frequency (76.9%) in T1 ESCC, and this gene was followed by NOTCH, MUC19, ZNF750, and FLG (mutation rates of 34.6%, 23.1%, 19.2%, and 15.3%, respectively). The recurrent focal copy number variants (CNVs) in tumor samples and paired noncancer samples from ESCC patients were analyzed by the SureCall copy number method after collecting comparative results of T1 ESCC vs CNVs from tumor tissues and paired noncancer tissues. Compared to that in noncancerous areas, the incidence of CNVs was significantly increased in the T1 ESCC region. In addition, the occurrence of CDKN2A deletion and CCND1 amplification was notably greater in the T1 ESCC region than in the noncancerous region (P < 0.001). Therefore, CNVs arise in the initial phases of cancer development, with CCND1 amplification and CDKN2A deletion playing pivotal roles in the initiation and advancement of ESCC.
The AJCC 8th edition is commonly used as a staging criterion for classifying ESCC into IA IB, IIA, IIB, IIIA, IIIB, IVA, and IVB. However, Li et al[26] divided samples collected from ESCC patients into three distinct phases, namely, nontumor, intraepithelial neoplasia (IEN), and advanced-stage ESCC (A-ESCC), to explore key mutational events in early-stage EC. The genes that were shown to be mutated in the IEN phase included PCDHB16, BOC, SYNE2, BCL9L, and STAG2. Analysis of somatic copy number alterations in the entire exome revealed that within the ESCC cohort of the TCGA database, chromosome 3q (chr3) exhibited the most significant amplification event, particularly during the IEN phase. Furthermore, during the early stages of IEN, there was a notable presence of important genes located at chr3, such as TP53, PIK3CA, and SOX2.
Transcriptomics refers to the examination of all RNA transcripts within a group of cells by high-throughput methodologies[27]. The transcriptome serves as a crucial bridge connecting the phenotypic characteristics of cells with the genetic aspects of tumor biology, encompassing all the information encoded by RNA transcripts derived from DNA. Thus, transcriptome adjustments reflect different cellular states, developmental stages, and regulatory mechanisms[28].
Tumor and paired noncancer tissues obtained from patients with ESCC in Iran were statistically analyzed by real-time fluorescence quantification and reverse transcription-PCR (RT-PCR). The majority of tumors overexpressing MAEL were stage I or stage II (P < 0.001), indicating that the level of MAEL was greater in primary stage (I/II) tumors. Therefore, early ESCC progression primarily triggers the activation of MAEL, while the expression level of MAEL decreases in advanced tumors. Furthermore, MAEL could serve as a reliable indicator for identifying patients with early ESCC in Iran[29].
Previous studies have shown that MEIS1, a constituent of the three-amino acid loop extension (TALE) HOX gene family, plays a negative regulatory role in early ESCC progression by inhibiting the cell cycle and cell proliferation, and its dysregulation may lead to functional alterations in different cell signaling pathways[30-32]. RT-PCR was used to explore the potential association between MEIS1 and crucial genes implicated in various cell signaling pathways during the progression of ESCC. The mRNA expression levels of MEIS1 and various marker genes (SIZN1, SALL4, and HEY1) were measured using quantitative real-time PCR (qRT-PCR) to investigate the correlation between genes and MEIS1 involved in different cell signaling pathways in ESCC. Additionally, pairwise correspondence was examined to identify any potential correlations. A notable association was observed between HEY1 and MEIS1 (P < 0.05) and between SIZN1 and MEIS1 (P < 0.05) in the initial stages of ESCC (stage I/II) compared with the later stages (III/IV)[33].
Alterations in metabolic processes and metabolic reprogramming are widely recognized as distinctive characteristics in the development of malignant tumors and represent revolutionary hallmarks of cancer[34]. Metabolomics is an omics approach that simultaneously quantifies an extensive number of chemical compounds present in biological organisms, employing corresponding technical methods to analyze extracts from urine, plasma, or tissue.
Therefore, metabolomics could generate clear biochemical information on disease mechanisms, thereby offering novel insights into cell signaling pathways affected by diseases and presenting new opportunities for the advancement of diagnostic indicators and therapeutics[35].
Blood samples were collected from healthy controls (HCs) individuals, patients with ESCC, and patients with esophageal squamous cell dysplasia (ESCD). Stages 0 and I were classified as early stage, stages II and III were considered intermediate stage, and stage IV was categorized as advanced stage. The plasma metabolic profiles were obtained by gas chromatography-mass spectrometry (GC/MS), and the potential diagnostic value of the various methods was evaluated by calculating receiver operating characteristic (ROC) curves and adjusted odds ratios (ORs). The HCs, ESCD, and ESCC groups exhibited 35, 46, and 9 distinct metabolites, respectively. Additionally, the early, intermediate, and late stages were characterized by 3, 6, and 4 unique metabolites, respectively. As ESCC progresses, the levels of cysteine and α-tocopherol increase gradually, and the level of aminomalonic acid increases. Therefore, α-tocopherol, cysteine, and aminomalonic acid may serve as markers for the early diagnosis of ESCC[36].
Previous studies have not only identified a single metabolite via metabolomics as an early marker for diagnosing ESCC but also established a diagnostic model of multiple metabolites to diagnose ESCC. Zhu et al[37] examined the plasma metabolomic profiles of patients with ESCC and HCs individuals via ultra-performance liquid chromatography-tandem quadruple time-of-flight mass spectrometry (UPLC-QTOF/MS) and identified 8 metabolic markers. A model for ESCC diagnosis based on 8 metabolic markers hypoxanthine, proline betaine, indole acrylic acid, inosine, 9-decanoylcarnitine, docosahexaenoic acid, lysophosphatidylethanolamine (LPE) (20:4) and lysophosphatidylcholine (LPC) (20:5), was constructed by machine learning. The area under the curve (AUC) of the ROC curve for this model was 0.991 [95% confidence interval (95%CI): 0.981-1.000]. Moreover, the sensitivity (SE) and specificity (SP) were 98.8% and 94.9%, respectively. In the validation set, an AUC of 0.965 (95%CI: 0.936-0.993) was observed, while the SE and SP were 88.3% and 88.9%, respectively. Further investigation revealed a correlation between the progression of ESCC and three biomarkers, namely, indole acrylic acid, LPE (20:4), and LPC (20:5).
Epigenetic modifications are thought to influence gene expression and cell phenotype rather than DNA sequence. Many known determinants of DNA methylation, histone tail modifications, and noncoding RNA expression constitute epigenetic modifications. Within the field of cancer, the disruption of epigenetic programming results in dysregulated cell division, impaired differentiation, and increased resistance to apoptosis[38]. Numerous studies have confirmed that alterations in the epigenome affect almost every step of tumor occurrence, progression, and treatment. Therefore, epigenetics has important implications for tumor exploration, diagnosis, delamination, and therapeutic intervention.
DNA methylation, a form of epigenetic modification, is indispensable for maintaining genome structure and controlling gene expression, and approximately 80% of CpG sequences in the human genome exhibit detectable DNA methylation[39]. Aberrant DNA methylation can contribute to the pathogenesis of malignancies. DNA hypermethylation can result in transcriptional repression and decreased gene expression, while DNA hypomethylation impacts chromosome stability and promotes aneuploidy[40,41]. Methylation of gene promoters that deviate from the norm may result in the inhibition of tumor suppressor genes, consequently impacting related transcriptional pathways and promoting tumor growth[42].
By analyzing the pathogenicity of DNA methylation in ESCC, it was found that the DNA methylation pattern gradually changes in the development and progression of EC, suggesting that it serves as an initial event during the progression of ESCC. The occurrence of methylation in the NKD2 gene is a common phenomenon observed in ESCC, and the methylation patterns of both the NKD2 and DACT2 genes are correlated with the classification of tumors according to TNM staging and lymph node metastasis. These results indicate that abnormal methylation patterns of DACT2 and NKD2 can be utilized as indicators for the timely identification and treatment of ESCC[43].
The amino terminal tail of histones is subject to a range of enzymatic modifications that induce diverse posttranslational alterations. The targeting of histone-modifying enzymes (HMEs) is responsible for chemical modifications of histones through covalent bonding, including methylation, acetylation, phosphorylation, and ubiquitination. These enzymes are pivotal for regulating both physiological and pathological biological processes[44]. The control of transcriptional activation and repression is suggested to involve the interaction of various HMEs, in addition to other epigenetic mechanisms, which collectively influence chromatin organization and subsequent gene expression[45].
Although there are many ways to modify histones, methylation and acetylation are the most common modifications. Chen et al[46] measured the quantities of the acetylated protein H3/H4 and the methylated protein H3K4/H3K27. This study revealed that the acetylation levels of histones H3 and H4 gradually decreased in tumor tissues. In addition, H3 exhibited reduced acetylation, while H3K27 methylation was significantly associated with tumor severity. In addition, hypoacetylation of H3 and excessive methylation of H3K4 and H3K27 were associated with the differentiation of tumor tissues. Therefore, we determined that tumor severity and tumor tissue differentiation were correlated with both H3K27 hypermethylation and H3 hypoacetylation. Additionally, the differentiation of tumor tissue was linked to H3K4 hypermethylation.
The acetylation/deacetylation of histones results from the regulatory oversight of two opposing enzymatic activities, namely, histone acetyltransferases (HATs) and histone deacetylases (HDACs)[47]. Since HDACs were first isolated, at least eight HDACs have been identified, and in the course of cancer development, HATs and HDACs play a role in modulating chromatin structure[48]. The acetylation status of histone H4 and the expression level of HDAC1 in human ESCC tissue were analyzed by Yasushitoh using immunohistochemistry. Histone H4 is exceedingly hyperacetylated in early ESCC, after which the protein is converted to a hypoacetylated state depending on the extent of carcinoma progression. This finding suggested that HAT activity is suppressed in advanced EC cells. The extent of acetylation on histone H4 and HDAC1 decreases correspondingly as cancer infiltrates the more profound strata of the esophageal wall. Moreover, it was observed that there is spatial similarity between high histone H4 acetylation and high HDAC1 expression within the same tumor[49]. Therefore, it can be speculated that the level of histone H4 acetylation and HDAC1 expression are important for early ESCC detection.
Noncoding RNAs (ncRNAs) play pivotal roles in human tumor formation and the pathogenesis and progression of malignant neoplasms. They function as regulators, either promoting or inhibiting the development and progression of carcinomas. NcRNAs are categorized according to their length, shape, and various locations. MicroRNAs (miRNAs), long-chain ncRNAs (lncRNAs), PIWI-interacting RNAs (piRNAs) and circular RNAs (circRNAs) constitute the primary categories of ncRNAs that perform different functions in cancer[50].
MiRNAs, which are small ncRNAs, are closely associated with the progression of carcinoma[51]. The identification of distinct miRNA profiles in human malignancies may aid in tumor diagnosis and cancer treatment[52-54]. A recent study showed that researchers have discovered a substantial presence of stable miRNAs in human serum, and the distribution of miRNA concentrations is altered in cancer patients[55].
It is convenient to obtain miRNAs from serum, plasma, and other body fluids, and these materials have good stability. Zhang et al[56] utilized Solexa sequencing analysis to identify the significant upregulation of 25 serum miRNAs in patients with ESCC compared with those in the normal group. The identification of seven serum miRNAs as biomarkers for ESCC was achieved through RT-qPCR analysis. To assess the effectiveness of the seven-member miRNA diagnostic model, researchers calculated the risk scoring function (RSF) for ESCC and control samples using a risk scoring formula. An RSF cutoff of 8.10 was used to correctly predict stage I or II disease in 85 of 106 patients (80.2%) and stage III or IV disease in 32 of 43 patients (74.4%). This study revealed that the serum miR-10b-3p concentration can serve as a novel diagnostic marker for ESCC. Hypomethylation of the miR-10b-3p promoter leads to overexpression of miR-10b-3p, thereby inhibiting FOXO3 expression and promoting the proliferation and metastasis of ESCC[57].
MiRNAs can be obtained not only from body fluids but also from tissues. Fujihara et al[58] obtained matched sets of primary ESCC tissue samples and corresponding neighboring healthy tissues from patients with a pathological diagnosis of stage I ESCC. Then, unsupervised hierarchical cluster analysis was used to determine the variation in gene expression patterns between ESCC tissues and neighboring healthy tissues, and 48 differentially expressed miRNAs were revealed. To confirm the findings of the miRNA analysis, scientists conducted qRT-PCR analysis of sets of tissues and both ESCC and normal esophageal cell lines. For this analysis, they selected the most differentially expressed miRNAs, miR-21-5p, miR-146b-5p, and miR-210-3p, from among the paired tissues. Further analysis revealed that miR-146b-5p and miR-21-5p were upregulated in tumors, while miR-210-3p expression levels were decreased compared to those in adjacent healthy tissues. In superficial ESCC tissues and neighboring healthy tissues, certain miRNA expression levels exhibited noticeable differences, indicating the significant diagnostic value of differentially expressed miRNAs for early ESCC.
NcRNAs have been widely studied, and lncRNAs have also received increased amounts of attention in recent years. Zhou et al[59] conducted a genome-wide screen of ESCC patients to identify potential biomarkers of lncRNAs. Then, they incorporated 6 LncRNA biomarkers to develop a diagnostic model called the MLMRP score. To assess the diagnostic accuracy of the model under these conditions, the expression levels of six lncRNAs were measured using an RT-qPCR assay and evaluated in different center databases and platform cohorts. Finally, 5 lncRNAs (LINC01082, PGM5-AS1, LINC03016, AP003548.1, and ADAMTS9-AS1) were considerably downregulated, and 1 lncRNA (MIR503HG) was considerably upregulated. The study further classified the samples into early-stage (stage I/II) and late-stage (stage III/IV) patients and then examined the diagnostic effectiveness of the model in the early stage. The model differentiated between early phase I and phase II ESCC patients in the different cohorts, with an AUC, SE, and SP above 0.90 in each cohort, revealing the possible significance of the model as a promising instrument for timely detection. A study showed that the lncRNA KLF3-AS1 can regulate KLF3 by binding to miR-185-5p and acting as a ceRNA of miR-185-5p. This results in the upregulation of KLF3 mRNA and protein expression and ultimately inhibits ESCC migration and invasion[60].
CircRNAs are novel ncRNAs that often serve as miRNA sponges to further regulate host gene expression and protein transport processes[61]. Li et al[62] reported that cir-ITCH can act as a miRNA sponge, leading to increased expression levels of ITCH and ubiquitin-mediated Dvl2 degradation. These effects thereby inhibit classical Wnt signaling and further inhibit the expression of the oncogene c-Myc, thereby preventing ESCC tumor development. Sang et al[63] demonstrated that ciRS-7, a miR-876-5p sponge, led to increased expression of the MAGE-A family and promoted the progression of ESCC.
Emerging bioinformatics has significantly contributed to contemporary medical research, with numerous databases containing a large number of samples, and machine learning is gradually being applied to clinical research. Yu et al[64] integrated ESCC patient information from the TCGA and GEO databases and divided the samples into early and late ESCC samples according to the patient's pathological disease stage. Then, 9 optimal lncRNA biomarkers were identified by machine learning, and a support vector machine (SVM) classifier was further constructed to differentiate between samples from patients with early-stage and advanced-stage ESCC. The SVM classifier successfully differentiated the progression stages of 160 ESCC patients in 179 samples, achieving a comprehensive precision rate of 89.39%. The SE, SP, negative predictive value (NPV) and positive predictive value (PPV) were 88.51%, 90.22%, 89.53%, and 89.25%, respectively, and the AUC was 0.91. Using samples from the TCGA database to verify the diagnostic ability of the 9 identified lncRNAs for predicting ESCC progression, the SVM classifier was able to accurately distinguish the stages of ESCC with a comprehensive precision of 90.14%. Similarly, the SP, SE, NPV, and PPV of the SVM classifier were 100.00%, 73.08%, 86.54% and 100.00%, respectively, and the AUC was 0.915. Thus, the SVM classifier based on 9 lncRNAs was demonstrated to have the potential to diagnose early ESCC.
Research into the microbiome is a dramatically progressing field in human cancer[65]. The microbiome composition varies across different types of tumors[66]. The oral and esophageal microbiomes have recently emerged as significant contributors to the progression of ESCC[67]. Chinese individuals with reduced microbial richness in the lower esophagus and decreased diversity of saliva microbes may exhibit greater likelihoods of developing esophageal squamous dysplasia and ESCC, respectively[68]. The researchers collected esophageal biopsy and swab samples from healthy people and patients with esophageal disease using healthy mean relative abundance (ARA%) as a baseline. They identified 109 microorganisms in the samples based on sequencing data and found 12 microorganisms with differential abundances between the normal samples and the ESCC samples. Of these, the ARA% of 8 microorganisms showed an upward trend, while the ARA% of the other 4 microorganisms showed a downward trend[69].
Fusobacterium nucleatum (F. nucleatum) is a bacterium that thrives in oxygen-deprived environments that was found to exist inside cells under certain conditions[70]. This microorganism is typically present intraorally, as well as in the urogenital and gastrointestinal tracts, including both the intestines and upper digestive system. Previous studies have provided evidence that F. nucleatum has a significant impact on promoting the proliferation and metastasis of diverse forms of carcinomas, including oral squamous cell carcinoma, pharyngeal squamous cell carcinoma, pancreatic cancer, colorectal cancer, and breast cancer[71-74]. Zhang et al[75] demonstrated an appreciably greater abundance of F. nucleatum in malignant tissues than in neighboring healthy tissues. Notably, predominant clustering of F. nucleatum within cancerous tissues was observed. Additionally, the expression of F. nucleatum in TNM stage III/IV ESCC tissues was significantly greater than that in TNM stage I/ II tissues.
The association between F. nucleatum and ESCC has been established; however, the underlying pathogenesis of this disease has not been determined. The promotion of ESCC cell proliferation by F. nucleatum might be mediated through the AHR/CYP1A1/AKT signaling pathway[76]. F. nucleatum infiltration into ESCC cells was demonstrated by Nomoto et al[77]; these authors also revealed that tumor advancement is facilitated by the activation of the NF-κB pathway, which is mediated through receptor-interacting protein kinase 2 (RIPK2) and nucleotide oligomerization domain 1. Research revealed a direct association between METTL3 expression and the abundance of F. nucleatum in ESCC tissues. In addition, overexpression of METTL3 enhanced the growth and motility of ESCC cells. One of the reasons for the increase in METTL3 transcription is infection by intracellular F. nucleatum. F. nucleatum infection leads to the activation of METTL3-associated m6A modifications that upregulate c-Myc expression. YTHDF1 recognizes methylated c-Myc mRNA and enhances its mRNA stability, ultimately leading to the upregulation of c-Myc expression to promote ESCC invasion and metastasis[78]. Ding et al[79] observed a greater abundance of F. nucleatum in ESCC tissues than in neighboring healthy tissues, and this elevated level of F. nucleatum was linked to a decreased rate of survival. Moreover, experiments have shown that F. nucleatum can invade ESCC cells by increasing putrescine production, disrupting the metabolism of polyamines, and facilitating the uncontrolled growth of malignant ESCC cells.
The term “early-stage esophageal cancer (EEC)” refers to mucosal (T1a) and submucosal (T1b) cancers, regardless of the presence of lymph node metastasis[80]. Lei et al[81] verified that the expression of F. nucleatum was greater in EEC tissues than in noncancerous tissues, and that F. nucleatum levels were increased with the development of EC, as determined by fluorescence in situ hybridization and RT-PCR. F. nucleatum accelerated the initial progression of ESCC by increasing the level of the interleukin IL-32/ PRTN3 complex, which in turn activated the PI3K/AKT signaling pathway (Figure 3). The DNA extracted from saliva samples by Wei et al[82] demonstrated that the predictive accuracy of Streptococcus salivarius (S. salivarius), F. nucleatum, and Porphyromonas gingivalis (P. gingivalis) individually and in combination for ESCC patients was 69.0%, 56.5%, 61.8%, and 76.4%, respectively. The corresponding sensitivity values at the critical threshold were 69.3%, 22.7%, 35.2%, and 86.4%, respectively. Similarly, the respective specificities were -78.4% for S. salivarius, -96.1% for F. nucleatum, and -90.2% for P. gingivalis, and for the combination of the three bacteria, it was -58.8%. These results suggest that microbes, whether alone or in combination, can function as prognostic indicators for ESCC.
A deeper understanding of the molecular mechanism underlying F. nucleatum expression in the initial stages of ESCC could lead to the discovery of new screening markers for early detection and therapeutic targets.
P. gingivalis is a prevalent gram-negative anaerobic bacterium closely related to periodontal disease. In addition to invading parts of the upper digestive tract, P. gingivalis infection also occurs in EC tissue. The infection of P. gingivalis in EC is considered a priority[83].
Although there is mounting evidence supporting the protumor properties of P. gingivalis in mouth and gastrointestinal system cancers, the molecular mechanisms underlying its role in ESCC have largely been elusive[84]. Therefore, elucidating the key molecular targets of P. gingivalis during the initiation and progression of ESCC will provide valuable insights for early diagnosis. Targeted therapy studies have demonstrated that colonization by P. gingivalis suppresses the expression of programmed cell death factor 4, resulting in the accumulation of cancer stem cells within ESCC cells and facilitating malignant progression[85]. Moreover, prolonged colonization by P. gingivalis results in the upregulation of the inflammatory cytokine glycogen synthetase kinase 3β, which facilitates mitochondrial oxidative phosphorylation (mtOXPHOS) and fosters malignant progression in ESCC[86]. Similarly, Gao et al[87] presented novel findings indicating that P. gingivalis enhances the expression of glycoprotein A repeat dominance (GARP), thereby activating the TGF-β/Smad signaling pathway. Additionally, the upregulated expression of GARP and activation of TGF-β were found to be partially reliant on P. gingivalis fimbriae (FimA) (Figure 3). The phosphorylation of Smad2/3 by P. gingivalis and the upregulation of GARP lead to unfavorable prognoses in patients with ESCC.
Among the serum antibodies against P. gingivalis, IgG and IgA showed potential as serum biomarkers for ESCC, with a sensitivity/specificity of 29.17%/96.90% and 52.10%/70.81%, respectively. Furthermore, the diagnostic efficacy of serum IgA antibodies against P. gingivalis surpassed that of IgG for detecting early-stage ESCC (54.54% vs 20.45%)[88]. The presence of P. gingivalis in saliva can serve as an indicator of ESCC. Chen et al[89] revealed that the concentration of P. gingivalis in the saliva of individuals diagnosed with ESCC was significantly increased compared to that in the saliva of healthy individuals in the control group (P < 0.05). When utilizing P. gingivalis as a diagnostic tool for ESCC, the calculated AUC was 0.599, with an SE of 62.2% and an SP of 70%. Patients diagnosed with early-stage ESCC can achieve curative outcomes through endoscopic resection (ER), including methods such as endoscopic mucosal removal, and endoscopic submucosal dissection (ESD)[90]. However, it should be noted that a subset of patients may experience metastasis or relapse following ER, regardless of the depth of tumor infiltration[91]. Research findings indicated that the abundance of P. gingivalis in tumors infiltrating the mucosal myometria and submucosa was greater than in lesions limited exclusively to the epithelium or lamina propria. Moreover, there were positive correlations between the presence of excessive P. gingivalis and invasion depth, post-ESD stenosis, and disease recurrence in the same area. Cox regression analyses with a single variable and multiple factors demonstrated that P. gingivalis independently predicted post-ESD recurrence and exhibited strong predictive capabilities for post-ESD local recurrence[92].
The development of more precise and efficient detection methods is expected through further research on the correlation between P. gingivalis and ESCC, and these methods are anticipated to have a crucial impact on the early screening and diagnosis of this malignancy.
The utilization of AI in digestive endoscopy can offer supplementary diagnosis, intelligent navigation, and precise treatment, thereby enhancing the efficiency and accuracy of medical interventions and improving patient outcomes.
The major pathogenic factors of EC have not been fully elucidated, leading to a dearth of effective interventions for this disease. Secondary prevention aims to achieve early diagnosis and treatment through screening and has emerged as a practical approach to mitigate the risk of advanced EC and its associated mortality[93]. The utilization of biomarker-based disease detection enhances recent advancements in nonendoscopic screening, thereby increasing the potential for improved accuracy and accessibility in detecting early ESCC. However, endoscopy remains the preferred screening method for ESCC[94].
The current presentations of early ESCC are distinguished by noticeable alterations observed during endoscopy, such as delicate mucosal conditions, localized erythema, erosions, patches, and nodules[95]. Conventional white-light imaging (WLI) has high accuracy in detecting ESCC, but it lacks sensitivity in identifying squamous cell dysplasia due to the subtle vascular changes that are only slightly different from those of normal squamous mucosa[96]. The use of innovative endoscopic methods, such as dye spray pigment endoscopy, virtual pigment endoscopy, confocal laser endoscopy, and volume laser endoscopy, can significantly enhance the diagnostic accuracy of white light endoscopy[97]. Texture with color-enhanced imaging is an innovative form of endoscopy with improved image quality that improves dark areas in WLI images, improves texture by highlighting subtle surface changes, and increases the contrast between the mucosa surrounding gastrointestinal lesions. This technique can aid in detecting ESCC based on color differences[98]. The Lugol iodine staining method remains the gold standard for detecting severe early ESCC. Lugol, an essential stain containing iodine, is effectively taken up by glycogen in typical esophageal squamous cells. However, tumor cells in the esophagus exhibit low glycogen content, resulting in decreased uptake of iodine from Lugol dye. This phenomenon is observed as areas devoid of Lugol staining and is commonly referred to as the “pink sign”[99]. Fluorescence molecular endoscopy enables the specific targeting and fluorescence highlighting of protein targets that are overexpressed in dysplasia and cancer. Zhao et al[100] recognized and confirmed GLUT1 as a potential target for imaging at the molecular level. Additionally, fluorescence imaging following the local application of 2-DG 800 CW has been demonstrated to effectively distinguish high-grade dysplasia and ESCC from low-grade dysplasia and normal esophageal tissue.
The minimally invasive therapeutic option for early ESCC is ER. The identification of tumor invasion depth can contribute to more informed decisions regarding ESCC treatment. The utilization of narrow band imaging (NBI) to magnify endoscopy results has been demonstrated to enhance the accuracy of invasion depth diagnosis in individuals suffering from superficial ESCC[101]. Previous studies have indicated that the accuracy and specificity of endoscopic ultrasound (EUS) surpasses those of NBI magnifying endoscopy (ME-NBI) in terms of accurately identifying the infiltration depth of ESCC[102]. However, Ishihara et al[103] reported that supplementing EUS with nonmagnification endoscopy (non-ME) or ME did not improve diagnostic accuracy in determining T1 ESCC invasion depth. Kobayashi et al[104] discovered that the utilization of linked color imaging combined with a light-emitting diode light source improves endoscopic assessment for determining the extent of invasion in ESCC through the observation and analysis of vessel diameter and vessel angle within inner capillary loops.
With advancements in medical imaging and biomarker technology, the application of endoscopy for diagnosing early ESCC is likely to increase. Moreover, the integration of molecular biology-based biomarker detection techniques with endoscopy holds promise for providing more comprehensive diagnostic information for early ESCC.
AI was first proposed in 1956 and can be defined as the capacity of machines to acquire knowledge and discern patterns and relationships from a sufficient number of representative examples. Thereafter, these data were used to make efficient informed decisions[105]. With the advancement of AI, its application in the diagnosis of gastrointestinal tumors has become increasingly prevalent[15,106-108]. Due to the varying levels of expertise among endoscopists, numerous cases have gone undetected. The implementation of AI can effectively address interobserver discrepancies and significantly improve the detection rate of ESCC[15].
Upper gastrointestinal endoscopy has a significant impact on the early detection of ESCC. However, achieving proficiency in identifying superficial lesions requires extensive and ongoing training for endoscopists. The deep learning computer-aided diagnosis (CAD) system for WLI and NBI modes, derived from the YOLO v5 algorithm, was configured by Meng et al[109]. The CAD system demonstrated excellent performance, with an AUC of 0.982 (95%CI: 0.969-0.994), an accuracy of 92.9% (95%CI: 89.5%-95.2%), an SE of 91.9% (95%CI: 87.4%-94.9%), and an SP of 94.7% (95%CI: 89.0%-97.0%). Furthermore, compared with their independent assessments alone, the CAD results significantly improved the precision exhibited by individuals without specialized expertise in endoscopy (88% vs 78%, P < 0.001). The AI diagnostic method by Shiroma et al[110] achieved a 100% correct diagnosis rate for ESCC (64/64) while also accurately detecting 85% of T1 ESCC patients (17/20). In comparison, WLI and NBI detected 75% (15/20) and 55% (11/22) of T1 ESCC patients, respectively. A study conducted by Wei et al[82] utilized a dataset of tens of thousands WLI images obtained from 1239 patients for the purpose of training and assessing an AI model based on a deep convolutional neural network. This model demonstrated high accuracy in identifying and outlining the boundaries of early ESCC lesions in datasets for both internal and external validation. A comparative analysis of WLI and nonmagnetic endoscopic narrowband imaging (NM-NBI) with a CAD system revealed that CAD-NBI exhibited greater specificity and accuracy than did CAD-WLI, while the sensitivity of CAD-WLI surpassed that of CAD-NBI. By utilizing a CAD system, endoscopists can enhance the diagnostic ability of these devices to achieve optimal levels of disease[111].
The ME-NBI technique is crucial for predicting the extent of invasion in ESCC patients due to its association with morphological changes observed in the intrapapillary capillary loop (IPCL) pattern[112]. After training and validation by Yuan et al[113] using a dataset of 685 ME-NBI images comprising 7094 patients, the internal and external validation cohorts yielded combined accuracies of 91.3% and 89.8%, respectively, as demonstrated by the AI system for diagnosing IPCL subtypes. With the aid of the AI system, leading endoscopists more accurately diagnosed both IPCL subtypes and depth of invasion. Zhang et al[114] developed an AI-based invasion depth prediction system (AI-IDPS) for ESCC utilizing invasion depth-related features. In terms of image verification, the AI-IDPS demonstrated a sensitivity, specificity, and accuracy of 85.7%, 86.3%, and 86.2%, respectively, in diagnosing SM2-3 lesions. For continuous video acquisition, the corresponding values were 87.5%, 84.0%, and 84.9%, respectively.
The utility of an AI system for real-time diagnosis of ESCC in a medical environment was assessed in a study involving 237 lesions. The accuracy, sensitivity and specificity of the AI systems were 80.6%, 68.2%, and 83.4%, respectively. In contrast, endoscopic physicians exhibited increased accuracy (85.7%) but decreased sensitivity (61.4%) and specificity (91.2%)[115]. Although there are still several limitations in the early detection of ESCC using AI, such as issues related to data quality, unreliable results, and the need for verification by professional doctors, significant improvements can be achieved by establishing comprehensive databases, enhancing model interpretability, integrating multimodal information, and strengthening self-learning capabilities. These advancements will further enhance the accuracy and reliability of AI in this field and provide better support for early diagnosis and treatment.
Endoscopy and AI technology each possess advantages in the early detection of ESCC. The combination of these two modalities can complement one another, enhancing diagnostic accuracy and efficiency.
With the deepening of research, multi-omics analysis has become an indispensable part of the study of disease progression. Through the combined analysis of multiple omics, the possible mechanisms, and therapeutic targets of ESCC were more fully revealed[57,62,63]. In the future, as endoscopic technology and AI continue to advance, there is vast potential for their integration in diagnosing early ESCC. Endoscopy provides visual lesion information[101,104], while AI technology offers more precise diagnostic results through extensive data analysis[88,111]. This amalgamation will provide a more accurate and convenient approach for diagnosing early ESCC.
The field of early detection of ESCC has undergone significant scientific advancements in recent years. Novel discoveries in biomarkers, molecular biology, epigenetics, microbiome research, endoscopy, and AI offer novel insights and methodologies for the timely identification of ESCC. Future studies should further explore these areas to improve the prognosis and quality of life of individuals diagnosed with ESCC.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Habu T, Japan S-Editor: Chen YL L-Editor: A P-Editor: Yu HG
| 1. | Wu HX, Pan YQ, He Y, Wang ZX, Guan WL, Chen YX, Yao YC, Shao NY, Xu RH, Wang F. Clinical Benefit of First-Line Programmed Death-1 Antibody Plus Chemotherapy in Low Programmed Cell Death Ligand 1-Expressing Esophageal Squamous Cell Carcinoma: A Post Hoc Analysis of JUPITER-06 and Meta-Analysis. J Clin Oncol. 2023;41:1735-1746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 2. | Thrift AP. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nat Rev Gastroenterol Hepatol. 2021;18:432-443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 120] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 3. | Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, Wang LD, Wang L, Yang W, Velculescu VE, Vogelstein B, Papadopoulos N, Kinzler KW, Meltzer SJ. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899-905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 313] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 4. | Li K, Lin Y, Zhou Y, Xiong X, Wang L, Li J, Zhou F, Guo Y, Chen S, Chen Y, Tang H, Qiu X, Cai S, Zhang D, Bremer E, Jim Yeung SC, Zhang H. Salivary Extracellular MicroRNAs for Early Detection and Prognostication of Esophageal Cancer: A Clinical Study. Gastroenterology. 2023;165:932-945.e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 5. | Li K, Lin Y, Luo Y, Xiong X, Wang L, Durante K, Li J, Zhou F, Guo Y, Chen S, Chen Y, Zhang D, Yeung SJ, Zhang H. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: a multicenter prospective study. Mol Cancer. 2022;21:21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 68] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 6. | Talukdar FR, di Pietro M, Secrier M, Moehler M, Goepfert K, Lima SSC, Pinto LFR, Hendricks D, Parker MI, Herceg Z. Molecular landscape of esophageal cancer: implications for early detection and personalized therapy. Ann N Y Acad Sci. 2018;1434:342-359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 7. | Jin X, Liu L, Wu J, Jin X, Yu G, Jia L, Wang F, Shi M, Lu H, Liu J, Liu D, Yang J, Li H, Ni Y, Luo Q, Jia W, Wang W, Chen WL. A multi-omics study delineates new molecular features and therapeutic targets for esophageal squamous cell carcinoma. Clin Transl Med. 2021;11:e538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2022;13:877-919. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 162] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 9. | Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016;375:2369-2379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1826] [Cited by in F6Publishing: 1904] [Article Influence: 238.0] [Reference Citation Analysis (0)] |
| 10. | Jiang Z, Wang J, Shen Z, Zhang Z, Wang S. Characterization of Esophageal Microbiota in Patients With Esophagitis and Esophageal Squamous Cell Carcinoma. Front Cell Infect Microbiol. 2021;11:774330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Adlung L, Elinav E, Greten TF, Korangy F. Microbiome genomics for cancer prediction. Nat Cancer. 2020;1:379-381. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Chu LY, Peng YH, Weng XF, Xie JJ, Xu YW. Blood-based biomarkers for early detection of esophageal squamous cell carcinoma. World J Gastroenterol. 2020;26:1708-1725. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 29] [Cited by in F6Publishing: 31] [Article Influence: 7.8] [Reference Citation Analysis (2)] |
| 13. | Chiang HC, Hughes M, Chang WL. The role of microbiota in esophageal squamous cell carcinoma: A review of the literature. Thorac Cancer. 2023;14:2821-2829. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 14. | Rai V, Abdo J, Agrawal DK. Biomarkers for Early Detection, Prognosis, and Therapeutics of Esophageal Cancers. Int J Mol Sci. 2023;24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 15. | Pan Y, He L, Chen W, Yang Y. The current state of artificial intelligence in endoscopic diagnosis of early esophageal squamous cell carcinoma. Front Oncol. 2023;13:1198941. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 16. | Liu W, Yuan X, Guo L, Pan F, Wu C, Sun Z, Tian F, Yuan C, Zhang W, Bai S, Feng J, Hu Y, Hu B. Artificial Intelligence for Detecting and Delineating Margins of Early ESCC Under WLI Endoscopy. Clin Transl Gastroenterol. 2022;13:e00433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 367] [Cited by in F6Publishing: 378] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 18. | Melhado RE, Alderson D, Tucker O. The changing face of esophageal cancer. Cancers (Basel). 2010;2:1379-1404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 19. | Siewert JR, Ott K. Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat Oncol. 2007;17:38-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 222] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 20. | Kamangar F, Chow WH, Abnet CC, Dawsey SM. Environmental causes of esophageal cancer. Gastroenterol Clin North Am. 2009;38:27-57, vii. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 406] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 21. | Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med. 2017;14:33-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 205] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 22. | Liu X, Wang X, Lin S, Yuan J, Yu IT. Dietary patterns and oesophageal squamous cell carcinoma: a systematic review and meta-analysis. Br J Cancer. 2014;110:2785-2795. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Maruvada P, Wang W, Wagner PD, Srivastava S. Biomarkers in molecular medicine: cancer detection and diagnosis. Biotechniques. 2005;Suppl: 9-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 24. | Chakravarthi BV, Nepal S, Varambally S. Genomic and Epigenomic Alterations in Cancer. Am J Pathol. 2016;186:1724-1735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 25. | Urabe Y, Kagemoto K, Hayes CN, Nakamura K, Masuda K, Ono A, Tanaka S, Arihiro K, Chayama K. Genomic characterization of early-stage esophageal squamous cell carcinoma in a Japanese population. Oncotarget. 2019;10:4139-4148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Li L, Jiang D, Zhang Q, Liu H, Xu F, Guo C, Qin Z, Wang H, Feng J, Liu Y, Chen W, Zhang X, Bai L, Tian S, Tan S, Xu C, Song Q, Zhong Y, Chen T, Zhou P, Zhao JY, Hou Y, Ding C. Integrative proteogenomic characterization of early esophageal cancer. Nat Commun. 2023;14:1666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 27. | Oberg JA, Glade Bender JL, Sulis ML, Pendrick D, Sireci AN, Hsiao SJ, Turk AT, Dela Cruz FS, Hibshoosh H, Remotti H, Zylber RJ, Pang J, Diolaiti D, Koval C, Andrews SJ, Garvin JH, Yamashiro DJ, Chung WK, Emerson SG, Nagy PL, Mansukhani MM, Kung AL. Implementation of next generation sequencing into pediatric hematology-oncology practice: moving beyond actionable alterations. Genome Med. 2016;8:133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 121] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 28. | Cieślik M, Chinnaiyan AM. Cancer transcriptome profiling at the juncture of clinical translation. Nat Rev Genet. 2018;19:93-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 152] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 29. | Abbaszadegan MR, Taghehchian N, Aarabi A, Akbari F, Saburi E, Moghbeli M. MAEL as a diagnostic marker for the early detection of esophageal squamous cell carcinoma. Diagn Pathol. 2021;16:36. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 30. | Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol. 2001;188:161-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 169] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 31. | Crist RC, Roth JJ, Waldman SA, Buchberg AM. A conserved tissue-specific homeodomain-less isoform of MEIS1 is downregulated in colorectal cancer. PLoS One. 2011;6:e23665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 32. | Song F, Wang H, Wang Y. Myeloid ecotropic viral integration site 1 inhibits cell proliferation, invasion or migration in human gastric cancer. Oncotarget. 2017;8:90050-90060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 33. | Mahmoudian RA, Forghanifard MM. Crosstalk between MEIS1 and markers of different cell signaling pathways in esophageal squamous cell carcinoma. Mol Biol Rep. 2020;47:3439-3448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297-308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2205] [Cited by in F6Publishing: 2292] [Article Influence: 191.0] [Reference Citation Analysis (0)] |
| 35. | Xu J, Cao W, Shao A, Yang M, Andoh V, Ge Q, Pan HW, Chen KP. Metabolomics of Esophageal Squamous Cell Carcinoma Tissues: Potential Biomarkers for Diagnosis and Promising Targets for Therapy. Biomed Res Int. 2022;2022:7819235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 36. | Yu M, Wen W, Yi X, Zhu W, Aa J, Wang G. Plasma Metabolomics Reveals Diagnostic Biomarkers and Risk Factors for Esophageal Squamous Cell Carcinoma. Front Oncol. 2022;12:829350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Zhu ZJ, Qi Z, Zhang J, Xue WH, Li LF, Shen ZB, Li ZY, Yuan YL, Wang WB, Zhao J. Untargeted Metabolomics Analysis of Esophageal Squamous Cell Carcinoma Discovers Dysregulated Metabolic Pathways and Potential Diagnostic Biomarkers. J Cancer. 2020;11:3944-3954. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 38. | Werner RJ, Kelly AD, Issa JJ. Epigenetics and Precision Oncology. Cancer J. 2017;23:262-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 39. | Craig JM, Bickmore WA. The distribution of CpG islands in mammalian chromosomes. Nat Genet. 1994;7:376-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 180] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 40. | Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489-492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1147] [Cited by in F6Publishing: 1072] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 41. | Min HL, Kim J, Kim WH, Jang BG, Kim MA. Epigenetic Silencing of the Putative Tumor Suppressor Gene GLDC (Glycine Dehydrogenase) in Gastric Carcinoma. Anticancer Res. 2016;36:179-187. [PubMed] [Cited in This Article: ] |
| 42. | Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18:1-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 235] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 43. | Ma K, Cao B, Guo M. The detective, prognostic, and predictive value of DNA methylation in human esophageal squamous cell carcinoma. Clin Epigenetics. 2016;8:43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 44. | Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381-395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3359] [Cited by in F6Publishing: 3483] [Article Influence: 267.9] [Reference Citation Analysis (0)] |
| 45. | Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 478] [Cited by in F6Publishing: 519] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 46. | Chen C, Zhao M, Yin N, He B, Wang B, Yuan Y, Yu F, Hu J, Yin B, Lu Q. Abnormal histone acetylation and methylation levels in esophageal squamous cell carcinomas. Cancer Invest. 2011;29:548-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 47. | Hassig CA, Schreiber SL. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300-308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 288] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 48. | Wang C, Fu M, Mani S, Wadler S, Senderowicz AM, Pestell RG. Histone acetylation and the cell-cycle in cancer. Front Biosci. 2001;6:D610-D629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 49. | Toh Y, Yamamoto M, Endo K, Ikeda Y, Baba H, Kohnoe S, Yonemasu H, Hachitanda Y, Okamura T, Sugimachi K. Histone H4 acetylation and histone deacetylase 1 expression in esophageal squamous cell carcinoma. Oncol Rep. 2003;10:333-338. [PubMed] [Cited in This Article: ] |
| 50. | Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65:625-639. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 176] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 51. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25833] [Cited by in F6Publishing: 26799] [Article Influence: 1340.0] [Reference Citation Analysis (0)] |
| 52. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5705] [Cited by in F6Publishing: 5849] [Article Influence: 324.9] [Reference Citation Analysis (0)] |
| 53. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5384] [Cited by in F6Publishing: 5479] [Article Influence: 304.4] [Reference Citation Analysis (0)] |
| 54. | Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M, Baba H. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915-1922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 220] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 55. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3218] [Cited by in F6Publishing: 3384] [Article Influence: 211.5] [Reference Citation Analysis (0)] |
| 56. | Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, Zhang Y, Li Y, Qiu H, Xing J, Liang Z, Ren B, Zen K, Zhang CY. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871-1879. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 251] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 57. | Lu YF, Yu JR, Yang Z, Zhu GX, Gao P, Wang H, Chen SY, Zhang J, Liu MY, Niu Y, Wei XM, Wang W, Ye FJ, Zhang LX, Zhao Y, Sun GG. Promoter hypomethylation mediated upregulation of MicroRNA-10b-3p targets FOXO3 to promote the progression of esophageal squamous cell carcinoma (ESCC). J Exp Clin Cancer Res. 2018;37:301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 58. | Fujihara S, Kobara H, Nishiyama N, Hirose K, Iwama H, Masaki T. MicroRNA Expression Profiles in Superficial Esophageal Squamous Cell Carcinoma before Endoscopic Submucosal Dissection: A Pilot Study. Int J Mol Sci. 2021;22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 59. | Zhou M, Bao S, Gong T, Wang Q, Sun J, Li J, Lu M, Sun W, Su J, Chen H, Liu Z. The transcriptional landscape and diagnostic potential of long non-coding RNAs in esophageal squamous cell carcinoma. Nat Commun. 2023;14:3799. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 60. | Liu JQ, Deng M, Xue NN, Li TX, Guo YX, Gao L, Zhao D, Fan RT. lncRNA KLF3-AS1 Suppresses Cell Migration and Invasion in ESCC by Impairing miR-185-5p-Targeted KLF3 Inhibition. Mol Ther Nucleic Acids. 2020;20:231-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 61. | Zhao X, Zhong Y, Wang X, Shen J, An W. Advances in Circular RNA and Its Applications. Int J Med Sci. 2022;19:975-985. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 52] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 62. | Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001-6013. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 449] [Cited by in F6Publishing: 547] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 63. | Sang M, Meng L, Sang Y, Liu S, Ding P, Ju Y, Liu F, Gu L, Lian Y, Li J, Wu Y, Zhang X, Shan B. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 2018;426:37-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 109] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 64. | Yu J, Wu X, Huang K, Zhu M, Zhang X, Zhang Y, Chen S, Xu X, Zhang Q. Bioinformatics identification of lncRNA biomarkers associated with the progression of esophageal squamous cell carcinoma. Mol Med Rep. 2019;19:5309-5320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 65. | Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, Sawayama H, Kinoshita K, Ishimoto T, Iwatsuki M, Sakamoto Y, Yamashita Y, Yoshida N, Watanabe M, Baba H. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin Cancer Res. 2016;22:5574-5581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 255] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 66. | Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, Meltser A, Douglas GM, Kamer I, Gopalakrishnan V, Dadosh T, Levin-Zaidman S, Avnet S, Atlan T, Cooper ZA, Arora R, Cogdill AP, Khan MAW, Ologun G, Bussi Y, Weinberger A, Lotan-Pompan M, Golani O, Perry G, Rokah M, Bahar-Shany K, Rozeman EA, Blank CU, Ronai A, Shaoul R, Amit A, Dorfman T, Kremer R, Cohen ZR, Harnof S, Siegal T, Yehuda-Shnaidman E, Gal-Yam EN, Shapira H, Baldini N, Langille MGI, Ben-Nun A, Kaufman B, Nissan A, Golan T, Dadiani M, Levanon K, Bar J, Yust-Katz S, Barshack I, Peeper DS, Raz DJ, Segal E, Wargo JA, Sandbank J, Shental N, Straussman R. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368:973-980. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 490] [Cited by in F6Publishing: 957] [Article Influence: 239.3] [Reference Citation Analysis (0)] |
| 67. | Yano Y, Etemadi A, Abnet CC. Microbiome and Cancers of the Esophagus: A Review. Microorganisms. 2021;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 68. | Abnet CC, Arnold M, Wei WQ. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2018;154:360-373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 935] [Cited by in F6Publishing: 930] [Article Influence: 155.0] [Reference Citation Analysis (0)] |
| 69. | Li M, Shao D, Zhou J, Gu J, Li X, Wei W. P1-13 Indicative microorganisms associated with the progression of esophageal squamous cell carcinoma (ESCC) in China. Ann Onco. 2021;32:S329. [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 70. | Li Y, Xing S, Chen F, Li Q, Dou S, Huang Y, An J, Liu W, Zhang G. Intracellular Fusobacterium nucleatum infection attenuates antitumor immunity in esophageal squamous cell carcinoma. Nat Commun. 2023;14:5788. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 71. | Lau HC, Yuan X, Huang H, Zhang M, Hsueh CY, Gong H. Fusobacterium nucleatum facilitates proliferation and autophagy by activating miR-361-3p/NUDT1 axis through oxidative stress in hypopharyngeal squamous cell carcinoma. BMC Cancer. 2023;23:990. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 72. | Ou S, Wang H, Tao Y, Luo K, Ye J, Ran S, Guan Z, Wang Y, Hu H, Huang R. Fusobacterium nucleatum and colorectal cancer: From phenomenon to mechanism. Front Cell Infect Microbiol. 2022;12:1020583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 12] [Reference Citation Analysis (0)] |
| 73. | Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, Yajuk O, Isaacson B, Abed J, Maalouf N, Nissan A, Sandbank J, Yehuda-Shnaidman E, Ponath F, Vogel J, Mandelboim O, Granot Z, Straussman R, Bachrach G. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. 2020;11:3259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 242] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 74. | Pignatelli P, Nuccio F, Piattelli A, Curia MC. The Role of Fusobacterium nucleatum in Oral and Colorectal Carcinogenesis. Microorganisms. 2023;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 9] [Reference Citation Analysis (0)] |
| 75. | Zhang JW, Zhang D, Yin HS, Zhang H, Hong KQ, Yuan JP, Yu BP. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression and chemoresistance by enhancing the secretion of chemotherapy-induced senescence-associated secretory phenotype via activation of DNA damage response pathway. Gut Microbes. 2023;15:2197836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 76. | Yin H, Zhang J, Zhang H, Li Q, Qiu H, Hong K, Wang W, Xiao Y, Yu B. Fusobacterium nucleatum promotes proliferation in oesophageal squamous cell carcinoma via AHR/CYP1A1 signalling. FEBS J. 2023;290:837-854. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 77. | Nomoto D, Baba Y, Liu Y, Tsutsuki H, Okadome K, Harada K, Ishimoto T, Iwatsuki M, Iwagami S, Miyamoto Y, Yoshida N, Watanabe M, Moroishi T, Komohara Y, Sawa T, Baba H. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression via the NOD1/RIPK2/NF-κB pathway. Cancer Lett. 2022;530:59-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 32] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 78. | Guo S, Chen F, Li L, Dou S, Li Q, Huang Y, Li Z, Liu W, Zhang G. Intracellular Fusobacterium nucleatum infection increases METTL3-mediated m6A methylation to promote the metastasis of esophageal squamous cell carcinoma. J Adv Res. 2023;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 79. | Ding N, Cheng Y, Liu H, Wu Y, Weng Y, Cui H, Cheng C, Zhang W, Cui Y. Fusobacterium nucleatum Infection Induces Malignant Proliferation of Esophageal Squamous Cell Carcinoma Cell by Putrescine Production. Microbiol Spectr. 2023;11:e0275922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 80. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 81. | Lei J, Xu F, Deng C, Nie X, Zhong L, Wu Z, Li J, Wu X, He S, Chen Y. Fusobacterium nucleatum promotes the early occurrence of esophageal cancer through upregulation of IL-32/PRTN3 expression. Cancer Sci. 2023;114:2414-2428. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 82. | Wei J, Li R, Lu Y, Meng F, Xian B, Lai X, Lin X, Deng Y, Yang D, Zhang H, Li L, Ben X, Qiao G, Liu W, Li Z. Salivary microbiota may predict the presence of esophageal squamous cell carcinoma. Genes Dis. 2022;9:1143-1151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 83. | Yuan X, Liu Y, Kong J, Gu B, Qi Y, Wang X, Sun M, Chen P, Sun W, Wang H, Zhou F, Gao S. Different frequencies of Porphyromonas gingivalis infection in cancers of the upper digestive tract. Cancer Lett. 2017;404:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 84. | Stasiewicz M, Karpiński TM. The oral microbiota and its role in carcinogenesis. Semin Cancer Biol. 2022;86:633-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 57] [Article Influence: 28.5] [Reference Citation Analysis (1)] |
| 85. | Li R, Liu Y, Zhou F, Yang H, Li J, Dai N, Sun W, Kong J, Gao S. Clinical Significance of Porphyromonas gingivalis Enriching Cancer Stem Cells by Inhibiting Programmed Cell Death Factor 4 in Esophageal Squamous Cell Carcinoma. ACS Infect Dis. 2023;9:1846-1857. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 86. | Liu Y, Zhou F, Yang H, Zhang Z, Zhang J, He K, Qian M, Li R, Sun W, Dai N, Li J, Guo Y, Kong J, Gao S. Porphyromonas gingivalis promotes malignancy and chemo-resistance via GSK3β-mediated mitochondrial oxidative phosphorylation in human esophageal squamous cell carcinoma. Transl Oncol. 2023;32:101656. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 87. | Gao S, Liu K, Jiao Y, Chen P, Gu B, Liu Y, Liang G, Shi L, Zhou F, Lamont RJ, Wang H, Qi YJ. Selective activation of TGFβ signaling by P. gingivalis-mediated upregulation of GARP aggravates esophageal squamous cell carcinoma. Am J Cancer Res. 2023;13:2013-2029. [PubMed] [Cited in This Article: ] |
| 88. | Gao SG, Yang JQ, Ma ZK, Yuan X, Zhao C, Wang GC, Wei H, Feng XS, Qi YJ. Preoperative serum immunoglobulin G and A antibodies to Porphyromonas gingivalis are potential serum biomarkers for the diagnosis and prognosis of esophageal squamous cell carcinoma. BMC Cancer. 2018;18:17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 89. | Chen X, Xian B, Wei J, Chen Y, Yang D, Lai X, Liu L, Wu Y, Lin X, Deng Y, Zhang H, Liu W, Qiao G, Li Z. Predictive value of the presence of Prevotella and the ratio of Porphyromonas gingivalis to Prevotella in saliva for esophageal squamous cell carcinoma. Front Cell Infect Microbiol. 2022;12:997333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 90. | Al-Haddad MA, Elhanafi SE, Forbes N, Thosani NC, Draganov PV, Othman MO, Ceppa EP, Kaul V, Feely MM, Sahin I, Ruan Y, Sadeghirad B, Morgan RL, Buxbaum JL, Calderwood AH, Chalhoub JM, Coelho-Prabhu N, Desai M, Fujii-Lau LL, Kohli DR, Kwon RS, Machicado JD, Marya NB, Pawa S, Ruan W, Sheth SG, Storm AC, Thiruvengadam NR, Qumseya BJ; (ASGE Standards of Practice Committee Chair). American Society for Gastrointestinal Endoscopy guideline on endoscopic submucosal dissection for the management of early esophageal and gastric cancers: methodology and review of evidence. Gastrointest Endosc. 2023;98:285-305.e38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 91. | Jiang D, Li X, Wang H, Xu C, Sujie A, Zeng H, Hou Y, Zhong Y. A retrospective study of endoscopic resection for 368 patients with early esophageal squamous cell carcinoma or precancerous lesions. Surg Endosc. 2017;31:2122-2130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 92. | Gao SG, Qi ZP, Qi YJ, Hou YY, Liu YW, Li MX, Li B, Sun D, Shi Q, Cai SL, Zhou PH, Zhong YS. Porphyromonas gingivalis predicts local recurrence after endoscopic submucosal dissection of early esophageal squamous cell carcinoma or precancerous lesion. BMC Cancer. 2023;23:43. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 93. | Li J, Xu J, Zheng Y, Gao Y, He S, Li H, Zou K, Li N, Tian J, Chen W, He J. Esophageal cancer: Epidemiology, risk factors and screening. Chin J Cancer Res. 2021;33:535-547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 53] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 94. | Codipilly DC, Qin Y, Dawsey SM, Kisiel J, Topazian M, Ahlquist D, Iyer PG. Screening for esophageal squamous cell carcinoma: recent advances. Gastrointest Endosc. 2018;88:413-426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 144] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 95. | Wen T, Wang W, Chen X. Recent advances in esophageal squamous cell precancerous conditions: A review. Medicine (Baltimore). 2022;101:e32192. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 96. | Wong MCS, Deng Y, Huang J, Bai Y, Wang HHX, Yuan J, Zhang L, Yip HC, Chiu PWY. Performance of screening tests for esophageal squamous cell carcinoma: a systematic review and meta-analysis. Gastrointest Endosc. 2022;96:197-207.e34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 97. | Visaggi P, Barberio B, Ghisa M, Ribolsi M, Savarino V, Fassan M, Valmasoni M, Marchi S, de Bortoli N, Savarino E. Modern Diagnosis of Early Esophageal Cancer: From Blood Biomarkers to Advanced Endoscopy and Artificial Intelligence. Cancers (Basel). 2021;13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 98. | Sugimoto M, Koyama Y, Itoi T, Kawai T. Using texture and colour enhancement imaging to evaluate gastrointestinal diseases in clinical practice: a review. Ann Med. 2022;54:3315-3332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 99. | Kandiah K, Chedgy FJ, Subramaniam S, Thayalasekaran S, Kurup A, Bhandari P. Early squamous neoplasia of the esophagus: The endoscopic approach to diagnosis and management. Saudi J Gastroenterol. 2017;23:75-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 100. | Zhao X, Huang Q, Koller M, Linssen MD, Hooghiemstra WTR, de Jongh SJ, van Vugt MATM, Fehrmann RSN, Li E, Nagengast WB. Identification and Validation of Esophageal Squamous Cell Carcinoma Targets for Fluorescence Molecular Endoscopy. Int J Mol Sci. 2021;22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 101. | Kim SJ, Kim GH, Lee MW, Jeon HK, Baek DH, Lee BE, Song GA. New magnifying endoscopic classification for superficial esophageal squamous cell carcinoma. World J Gastroenterol. 2017;23:4416-4421. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 25] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 102. | Suzuki Y, Nomura K, Kikuchi D, Iizuka T, Koseki M, Kawai Y, Okamura T, Ochiai Y, Hayasaka J, Mitsunaga Y, Odagiri H, Yamashita S, Matsui A, Ohashi K, Hoteya S. Diagnostic Performance of Endoscopic Ultrasonography with Water-Filled Balloon Method for Superficial Esophageal Squamous Cell Carcinoma. Dig Dis Sci. 2023;68:3974-3984. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 103. | Ishihara R, Mizusawa J, Kushima R, Matsuura N, Yano T, Kataoka T, Fukuda H, Hanaoka N, Yoshio T, Abe S, Yamamoto Y, Nagata S, Ono H, Tamaoki M, Yoshida N, Takizawa K, Muto M. Assessment of the Diagnostic Performance of Endoscopic Ultrasonography After Conventional Endoscopy for the Evaluation of Esophageal Squamous Cell Carcinoma Invasion Depth. JAMA Netw Open. 2021;4:e2125317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 104. | Kobayashi K, Miyahara R, Funasaka K, Furukawa K, Sawada T, Maeda K, Yamamura T, Ishikawa T, Ohno E, Nakamura M, Kawashima H, Nakaguro M, Okumura Y, Hirooka Y, Fujishiro M. Color information from linked color imaging is associated with invasion depth and vascular diameter in superficial esophageal squamous cell carcinoma. Dig Endosc. 2020;32:65-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 105. | Bhinder B, Gilvary C, Madhukar NS, Elemento O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021;11:900-915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 144] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 106. | Kenner B, Chari ST, Kelsen D, Klimstra DS, Pandol SJ, Rosenthal M, Rustgi AK, Taylor JA, Yala A, Abul-Husn N, Andersen DK, Bernstein D, Brunak S, Canto MI, Eldar YC, Fishman EK, Fleshman J, Go VLW, Holt JM, Field B, Goldberg A, Hoos W, Iacobuzio-Donahue C, Li D, Lidgard G, Maitra A, Matrisian LM, Poblete S, Rothschild L, Sander C, Schwartz LH, Shalit U, Srivastava S, Wolpin B. Artificial Intelligence and Early Detection of Pancreatic Cancer: 2020 Summative Review. Pancreas. 2021;50:251-279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 49] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 107. | Mitsala A, Tsalikidis C, Pitiakoudis M, Simopoulos C, Tsaroucha AK. Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era. Curr Oncol. 2021;28:1581-1607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 62] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 108. | Niu PH, Zhao LL, Wu HL, Zhao DB, Chen YT. Artificial intelligence in gastric cancer: Application and future perspectives. World J Gastroenterol. 2020;26:5408-5419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 56] [Cited by in F6Publishing: 51] [Article Influence: 12.8] [Reference Citation Analysis (1)] |
| 109. | Meng QQ, Gao Y, Lin H, Wang TJ, Zhang YR, Feng J, Li ZS, Xin L, Wang LW. Application of an artificial intelligence system for endoscopic diagnosis of superficial esophageal squamous cell carcinoma. World J Gastroenterol. 2022;28:5483-5493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 2] [Cited by in F6Publishing: 4] [Article Influence: 2.0] [Reference Citation Analysis (2)] |
| 110. | Shiroma S, Yoshio T, Kato Y, Horie Y, Namikawa K, Tokai Y, Yoshimizu S, Yoshizawa N, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Akazawa N, Akiyama J, Tada T, Fujisaki J. Ability of artificial intelligence to detect T1 esophageal squamous cell carcinoma from endoscopic videos and the effects of real-time assistance. Sci Rep. 2021;11:7759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 111. | Li B, Cai SL, Tan WM, Li JC, Yalikong A, Feng XS, Yu HH, Lu PX, Feng Z, Yao LQ, Zhou PH, Yan B, Zhong YS. Comparative study on artificial intelligence systems for detecting early esophageal squamous cell carcinoma between narrow-band and white-light imaging. World J Gastroenterol. 2021;27:281-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 112. | Oyama T, Inoue H, Arima M, Momma K, Omori T, Ishihara R, Hirasawa D, Takeuchi M, Tomori A, Goda K. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: magnifying endoscopic classification of the Japan Esophageal Society. Esophagus. 2017;14:105-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 186] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 113. | Yuan XL, Liu W, Liu Y, Zeng XH, Mou Y, Wu CC, Ye LS, Zhang YH, He L, Feng J, Zhang WH, Wang J, Chen X, Hu YX, Zhang KH, Hu B. Artificial intelligence for diagnosing microvessels of precancerous lesions and superficial esophageal squamous cell carcinomas: a multicenter study. Surg Endosc. 2022;36:8651-8662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 114. | Zhang L, Luo R, Tang D, Zhang J, Su Y, Mao X, Ye L, Yao L, Zhou W, Zhou J, Lu Z, Zhang M, Xu Y, Deng Y, Huang X, He C, Xiao Y, Wang J, Wu L, Li J, Zou X, Yu H. Human-Like Artificial Intelligent System for Predicting Invasion Depth of Esophageal Squamous Cell Carcinoma Using Magnifying Narrow-Band Imaging Endoscopy: A Retrospective Multicenter Study. Clin Transl Gastroenterol. 2023;14:e00606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 115. | Tani Y, Ishihara R, Inoue T, Okubo Y, Kawakami Y, Matsueda K, Miyake M, Yoshii S, Shichijo S, Kanesaka T, Yamamoto S, Takeuchi Y, Higashino K, Uedo N, Michida T, Kato Y, Tada T. A single-center prospective study evaluating the usefulness of artificial intelligence for the diagnosis of esophageal squamous cell carcinoma in a real-time setting. BMC Gastroenterol. 2023;23:184. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |