Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Dec 15, 2024; 16(12): 4700-4715
Published online Dec 15, 2024. doi: 10.4251/wjgo.v16.i12.4700
Published online Dec 15, 2024. doi: 10.4251/wjgo.v16.i12.4700
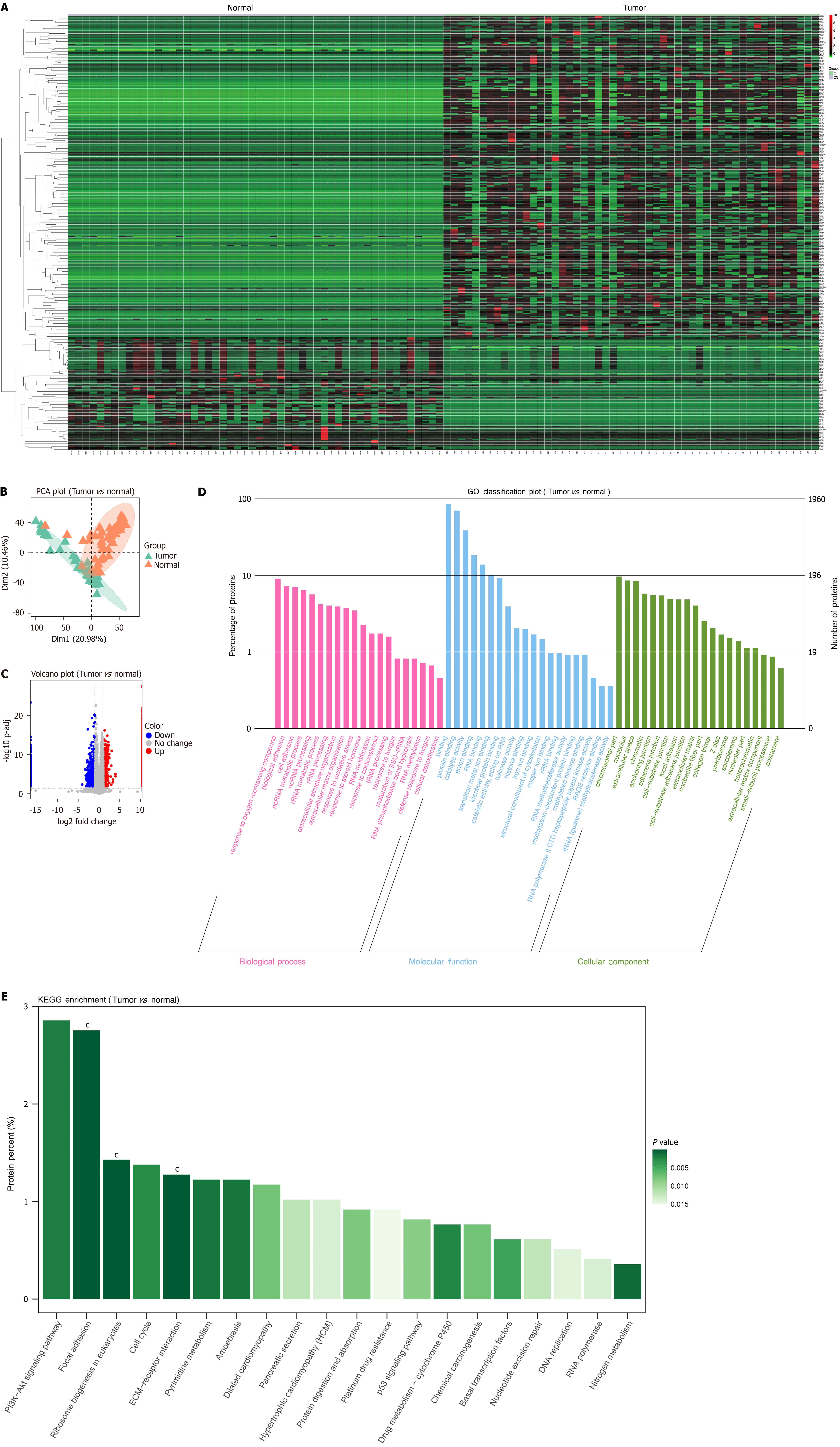
Figure 2 Proteomic profiles and bioinformatic analysis of colorectal cancer patients.
A: Hierarchical cluster analysis shows the global differentially expressed proteins. Upregulated (red) and downregulated (green) differentially expressed proteins in the CRC group compared to the normal group; B: Principal component analysis shows the protein expression in the cancer tissues (green) and normal tissues (orange) of colorectal cancer (CRC) patients; C: Volcano plot shows the fold changes of identified proteins. Significantly upregulated or downregulated proteins are represented in red and blue, respectively, while proteins without significant changes are shown in gray; D: Gene Ontology analysis shows the enrichment results for differentially expressed proteins; E: Kyoto encyclopedia of genes and genomes analysis shows that differentially expressed proteins are significantly enriched in multiple pathways, cP < 0.001. CRC: Colorectal cancer; PCA: Principal component analysis; HCA: Hierarchical cluster analysis; GO: Gene ontology; KEGG: Kyoto encyclopedia of genes and genomes; PI3K-Akt: Phosphatidylinositol 3-kinase serine/threonine kinase; ECM: Extracellular matrix.
- Citation: Zhou YC, Wang QQ, Zhou GYJ, Yin TF, Zhao DY, Sun XZ, Tan C, Zhou L, Yao SK. Matrine promotes colorectal cancer apoptosis by downregulating shank-associated RH domain interactor expression. World J Gastrointest Oncol 2024; 16(12): 4700-4715
- URL: https://www.wjgnet.com/1948-5204/full/v16/i12/4700.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i12.4700