Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Jan 15, 2024; 16(1): 30-50
Published online Jan 15, 2024. doi: 10.4251/wjgo.v16.i1.30
Published online Jan 15, 2024. doi: 10.4251/wjgo.v16.i1.30
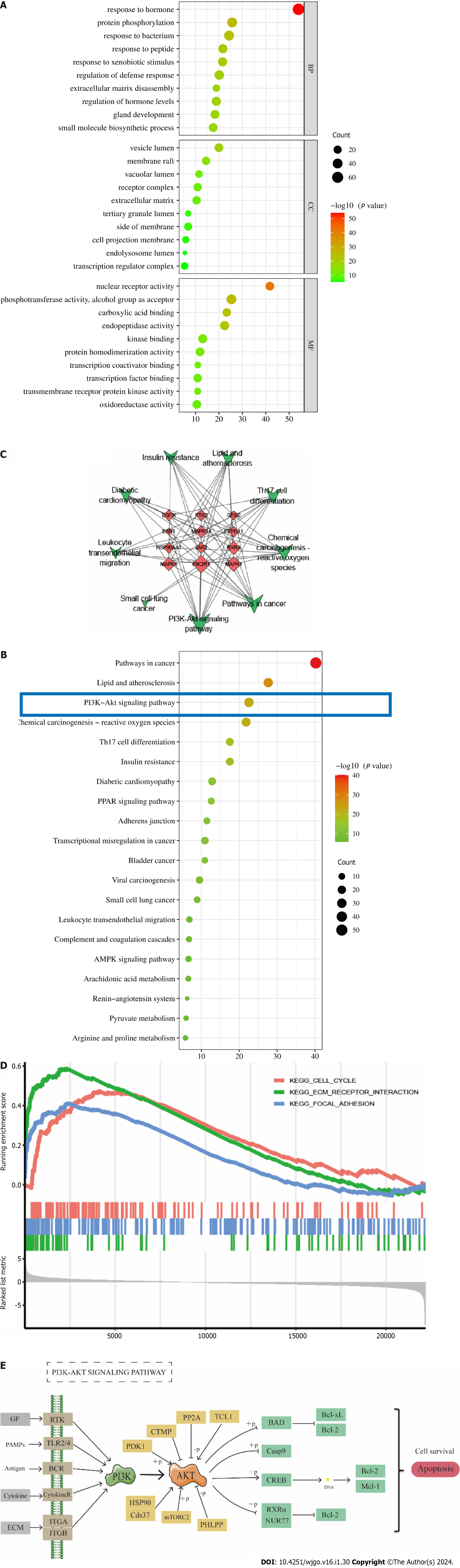
Figure 3 Enrichment analysis.
A: Gene Ontology enrichment analysis; B: Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis; C: Protein-protein interaction network diagram of KEGG pathways and top 12 core targets, green represents pathways and red represents core targets; D: Gene set enrichment analysis analyzed the results; E: PI3K/AKT signaling pathway. GF: Growth factor; PAMP: Pathogen-associated molecular patterns; ECM: von Willebrand factor; TLR2/4: Toll-like receptor 2/4; BCR: Breakpoint cluster region protein; ITGA: Integrin alpha; ITGB: Integrin beta; PDK1: Pyruvate dehydrogenase kinase 1; PP2A: Serine/threonine-protein phosphatase 2A catalytic subunit; TCL1: T-cell leukemia/lymphoma protein 1A; HSP90: Heat shock protein 90; mTORC2: CREB-regulated transcription coactivator 2; PHLPP: PH domain leucine-rich repeat-containing protein phosphatase 1; BAD: Bcl2-associated agonist of cell death; CREB: Ubiquitin carboxyl-terminal hydrolase creB.
- Citation: Du YH, Zhao JJ, Li X, Huang SC, Ning N, Chen GQ, Yang Y, Nan Y, Yuan L. Mechanism of pachymic acid in the treatment of gastric cancer based on network pharmacology and experimental verification. World J Gastrointest Oncol 2024; 16(1): 30-50
- URL: https://www.wjgnet.com/1948-5204/full/v16/i1/30.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i1.30