Copyright
©The Author(s) 2023.
World J Gastrointest Oncol. Aug 15, 2023; 15(8): 1384-1399
Published online Aug 15, 2023. doi: 10.4251/wjgo.v15.i8.1384
Published online Aug 15, 2023. doi: 10.4251/wjgo.v15.i8.1384
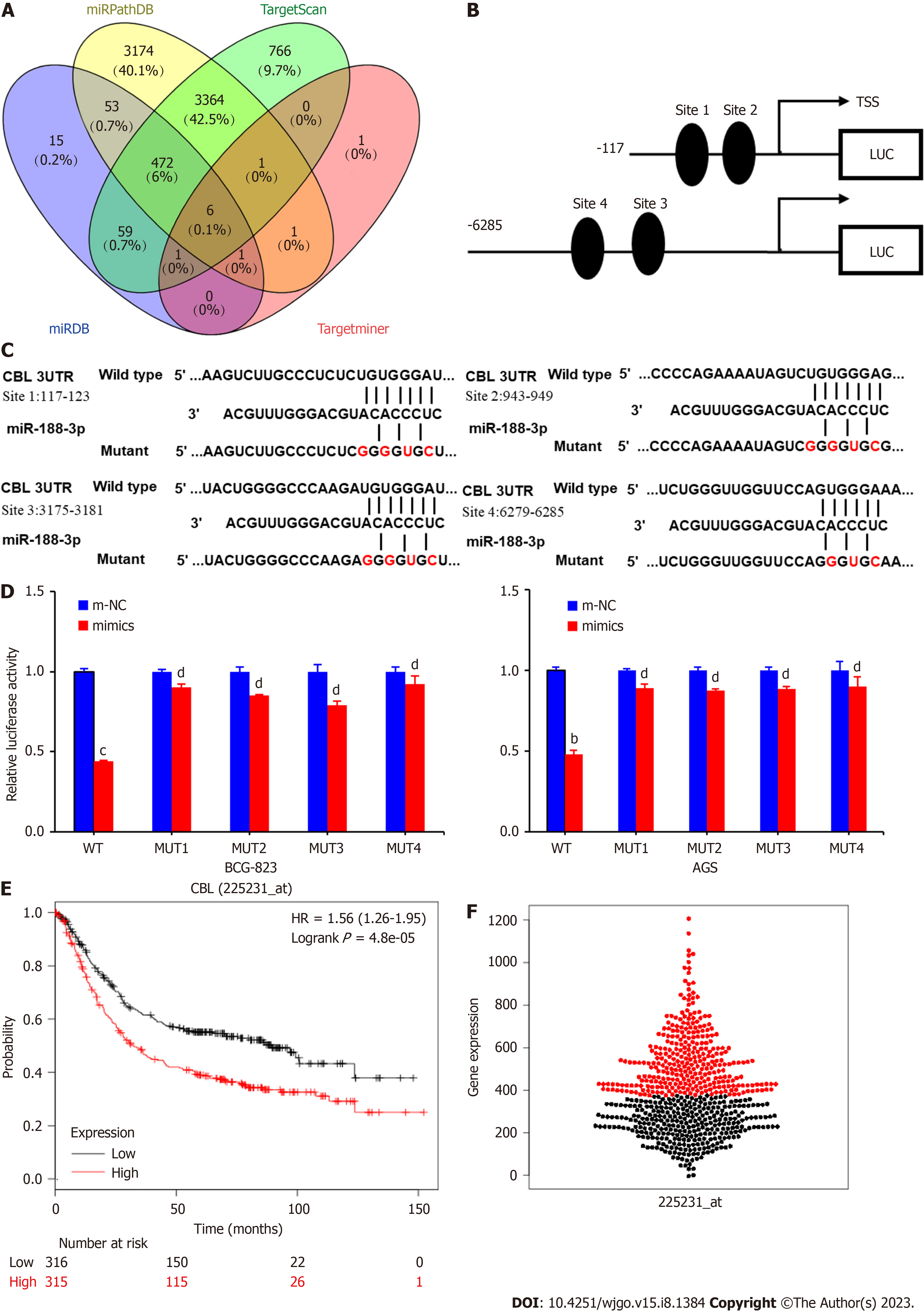
Figure 4 CBL as a direct target of miR-188-3p.
A: Bioinformatic prediction of miR-188-3p gene targets. We searched the TargetScan, miRPathDB, miRDB and TargetMiner databases for possible target genes of miR-188-3p. We selected six genes and CBL (an oncogene in gastric cancer) for a confirmation study; B and C: The putative miR-188-3p-binding sites of the CBL 3′-UTR. nt, nucleotides; D: Luciferase gene reporter assay. AGS cells were seeded in six-well plates for transient transfection with miR-188-3p mimics and with the reporter plasmid containing a luciferase gene fused to the target sequence of the wild-type (WT) 3'-UTR or mutant 3’-UTR of CBL using Lipofectamine 3000 for 36 h. Tumor cells were then lysed and an equal amount of protein samples was analyzed using the Luciferase Reporter Assay System. wt, wild type; mut, mutant; site 1, the miR-188-3p motif spanning nt:117-123 nt; site 2, the motif spanning nt 943-949 nt in the CBL 3′-UTR; site 3, the motif spanning nt 3175-3181nt in the CBL 3′-UTR; site 4, the motif spanning nt 6279-6285 nt in the CBL 3′-UTR. cP < 0.001, bP < 0.01, and dP > 0.05; E and F: Kaplan-Meier survival analysis. We utilized data from the KM-Plotter database (http://kmplot.com) and TCGA dataset (http://xena.ucsc.edu/public), and performed overall survival analysis.
- Citation: Lin JJ, Luo BH, Su T, Yang Q, Zhang QF, Dai WY, Liu Y, Xiang L. Antitumor activity of miR-188-3p in gastric cancer is achieved by targeting CBL expression and inactivating the AKT/mTOR signaling. World J Gastrointest Oncol 2023; 15(8): 1384-1399
- URL: https://www.wjgnet.com/1948-5204/full/v15/i8/1384.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i8.1384