Copyright
©The Author(s) 2023.
World J Gastrointest Oncol. Jul 15, 2023; 15(7): 1227-1240
Published online Jul 15, 2023. doi: 10.4251/wjgo.v15.i7.1227
Published online Jul 15, 2023. doi: 10.4251/wjgo.v15.i7.1227
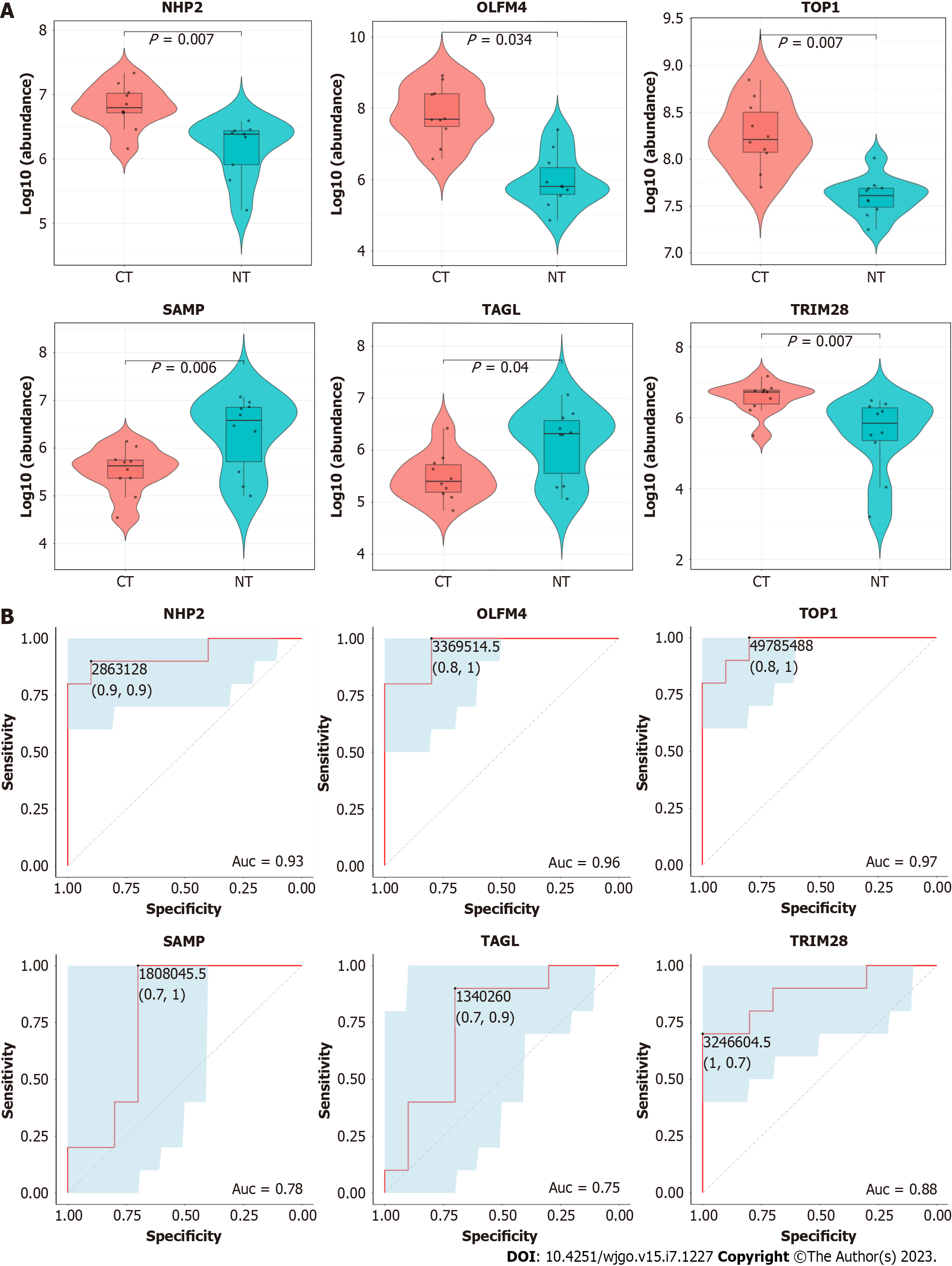
Figure 3 Verification of candidate exosomal proteins.
A: Parallel reaction monitoring analysis shows the upregulation of NHP2 (P = 0.007), OLFM4 (P = 0.034), TOP1 (P = 0.007), TRIM28 (P = 0.007), and the downregulation of SAMP (P = 0.006), TAGL(p=0.04) between colorectal cancer tissue exosomes and normal intestinal tissue exosomes; B: Receiver operating characteristic curve analysis shows that the area under curve of NHP2, OLFM4, TOP1, SAMP, TAGL, and TRIM28 are 0.93, 0.96, 0.97, 0.78, 0.75, 0.88, respectively (P < 0.05). CT: Colorectal cancer tissues; NT: Normal intestinal tissues; ROC: Receiver operating characteristic curve; AUC: Area under ROC curve; NHP2: H/ACA ribonucleoprotein complex subunit 2; OLFM4: Olfactomedin-4; TOP1: DNA topoisomerase 1; SAMP: Serum amyloid P-component; TAGL: Transgelin; TRIM28: Tripartite motif-containing protein 28.
- Citation: Zhou GYJ, Zhao DY, Yin TF, Wang QQ, Zhou YC, Yao SK. Proteomics-based identification of proteins in tumor-derived exosomes as candidate biomarkers for colorectal cancer. World J Gastrointest Oncol 2023; 15(7): 1227-1240
- URL: https://www.wjgnet.com/1948-5204/full/v15/i7/1227.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i7.1227