Copyright
©The Author(s) 2023.
World J Gastrointest Oncol. May 15, 2023; 15(5): 787-809
Published online May 15, 2023. doi: 10.4251/wjgo.v15.i5.787
Published online May 15, 2023. doi: 10.4251/wjgo.v15.i5.787
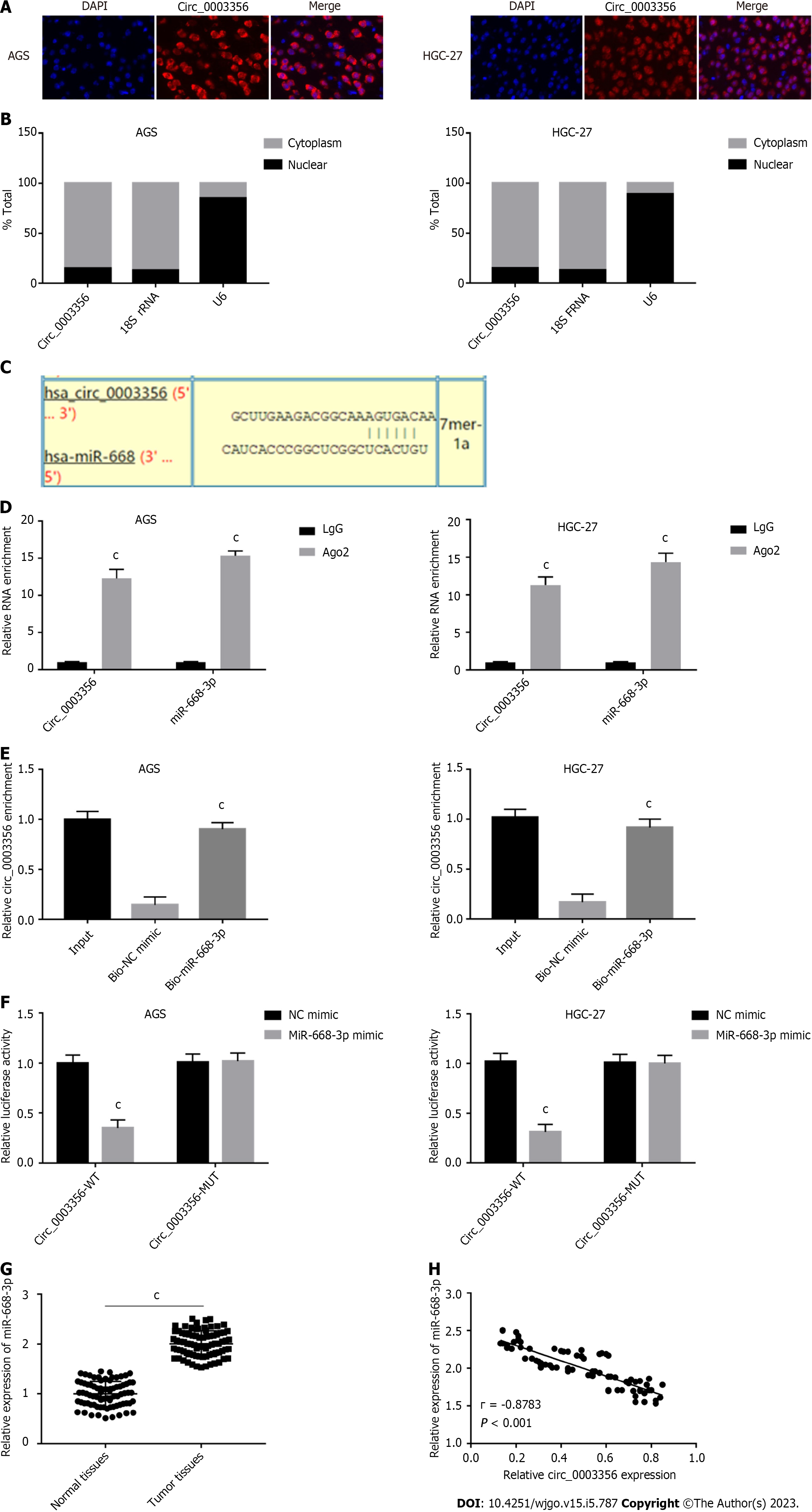
Figure 3 Circ_0003356 directly binds to miR-668-3p.
A: Fluorescence in situ hybridization assays were used to detect the sub-cellular localization of circ_0003356 in AGS and HGC-27 cells; B: Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the expression of circ_0003356 in the nucleus and cytoplasm; C: Binding sequences between circ_0003356 and miR-668-3p predicted by circinteractome; D: Interaction between circ_0003356 and miR-668-3p in AGS and HGC-27 cells validated by RIP-qRT-PCR; E: interaction between circ_0003356 and miR-668-3p in AGS and HGC-27 cells validated by RNA pull-down assays; F: Interaction between circ_0003356 and miR-668-3p in AGS and HGC-27 cells validated by dual-luciferase reporter assays; G: Relative expression of miR-668-3p detected by qRT-PCR in tumor tissues and normal tissues; H: Correlation between circ_0003356 and miR-668-3p in gastric cancer tissues analyzed by Pearson’s correlation analysis. cP < 0.001.
- Citation: Li WD, Wang HT, Huang YM, Cheng BH, Xiang LJ, Zhou XH, Deng QY, Guo ZG, Yang ZF, Guan ZF, Wang Y. Circ_0003356 suppresses gastric cancer growth through targeting the miR-668-3p/SOCS3 axis. World J Gastrointest Oncol 2023; 15(5): 787-809
- URL: https://www.wjgnet.com/1948-5204/full/v15/i5/787.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i5.787