Copyright
©The Author(s) 2023.
World J Gastrointest Oncol. Mar 15, 2023; 15(3): 372-388
Published online Mar 15, 2023. doi: 10.4251/wjgo.v15.i3.372
Published online Mar 15, 2023. doi: 10.4251/wjgo.v15.i3.372
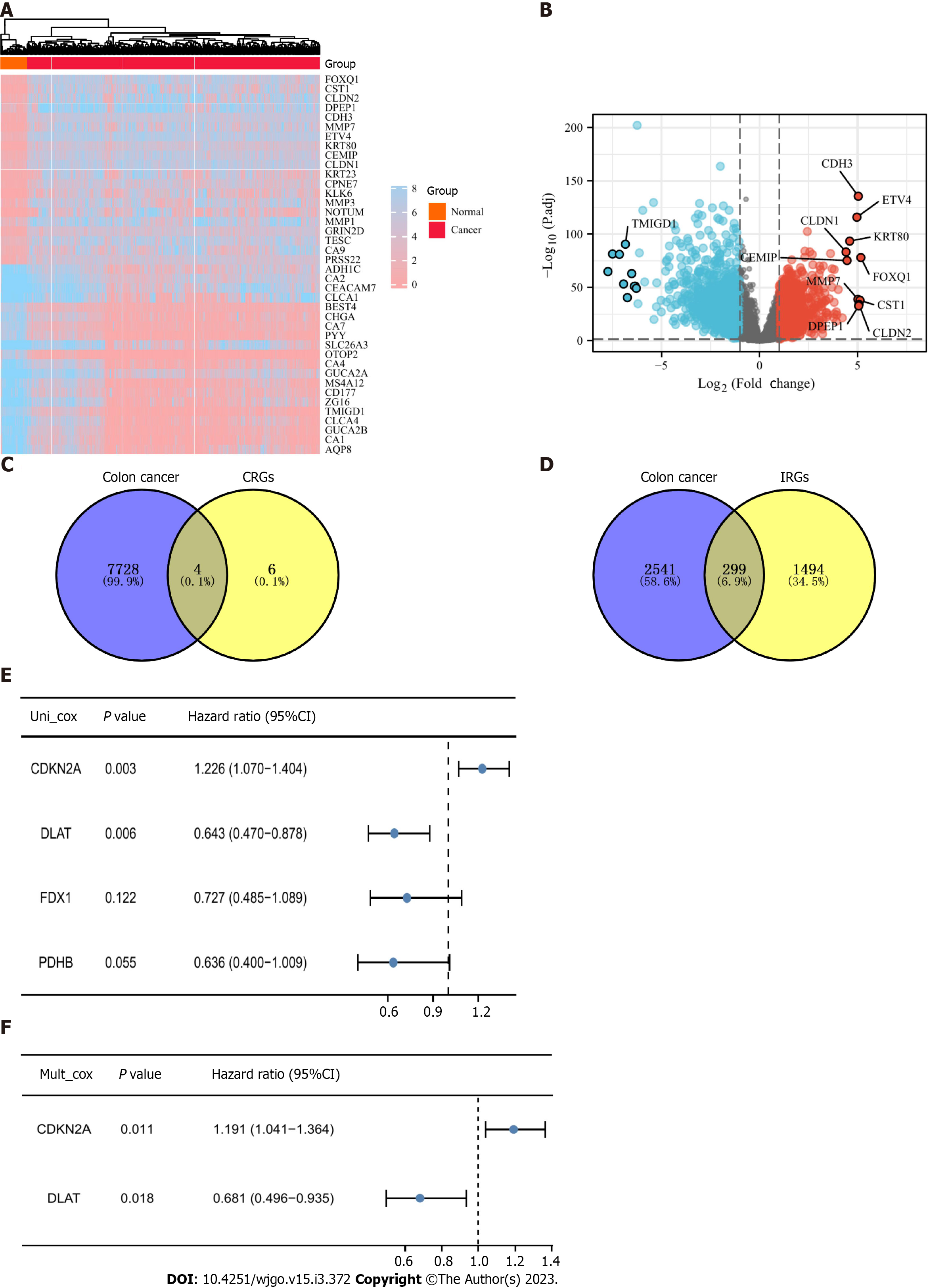
Figure 2 Acquisition of cuproptosis-related and immune-related differentially expressed genes.
A: Heatmap of differentially expressed genes in The Cancer Genome Atlas colon cancer patients compared with normal controls; B: Volcano plot of differentially expressed genes (DEGs), where blue represents downregulated genes and red represents upregulated genes; C: Four cuproptosis-related DEGs (CR-DEGs) were calculated by taking the intersection of DEGs (fold change > 1.2 and P.adjust < 0.05) and CRGs; D: DEGs with |log2FC| > 1 and p.adjust < 0.05 were intersected with immune-related genes (IRGs) to obtain IR-DEGs; E: Result of univariate Cox regression analysis of CR-DEGs; F: The result of multivariate Cox regression analysis of CR-DEGs.
- Citation: Huang YY, Bao TY, Huang XQ, Lan QW, Huang ZM, Chen YH, Hu ZD, Guo XG. Machine learning algorithm to construct cuproptosis- and immune-related prognosis prediction model for colon cancer. World J Gastrointest Oncol 2023; 15(3): 372-388
- URL: https://www.wjgnet.com/1948-5204/full/v15/i3/372.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i3.372