Copyright
©The Author(s) 2023.
World J Gastrointest Oncol. Jan 15, 2023; 15(1): 76-89
Published online Jan 15, 2023. doi: 10.4251/wjgo.v15.i1.76
Published online Jan 15, 2023. doi: 10.4251/wjgo.v15.i1.76
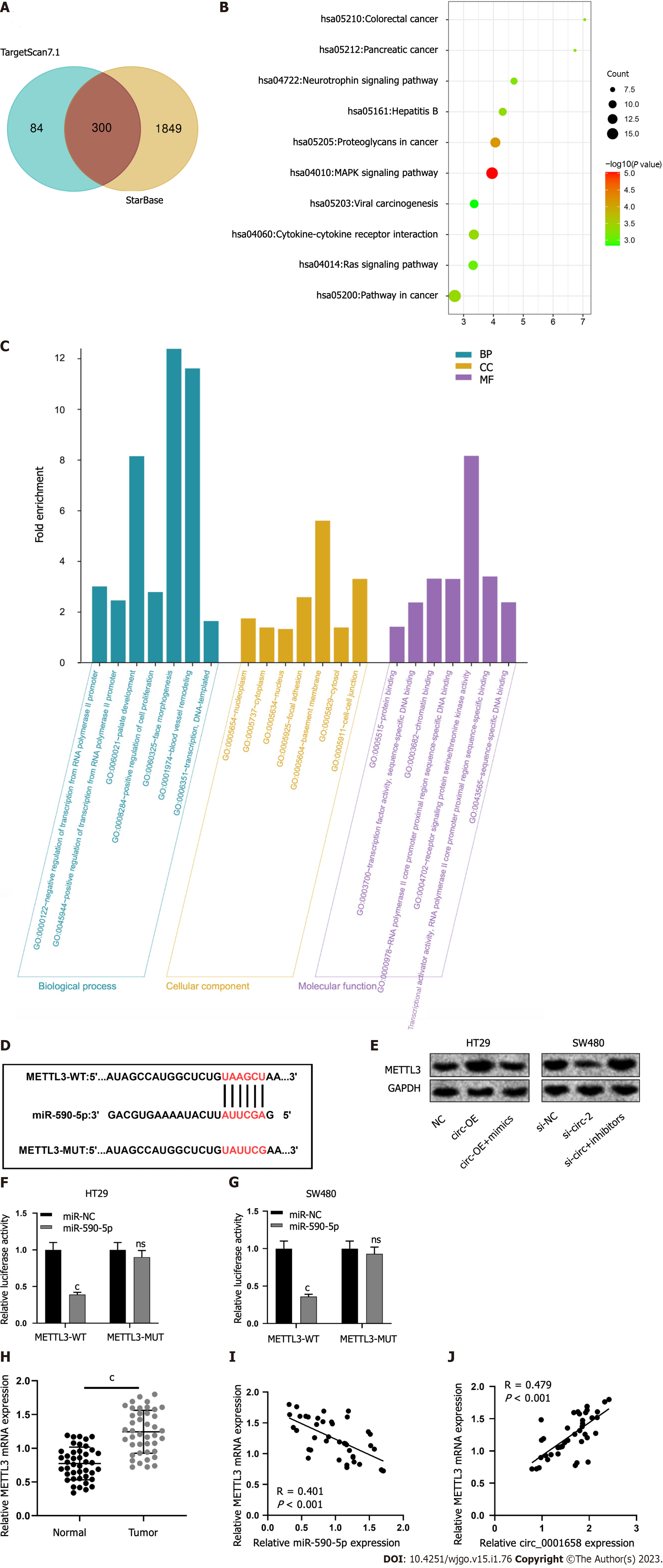
Figure 5 Circ_0001658 raises METTL3 expression by adsorption of miR-590-5p.
A: The downstream targets of miR-590-5p were under the prediction of the bioinformatics websites StarBase and TargetScan; B and C: Kyoto Encyclopedia of Genes and Genomes and Gene Ontology enrichment analysis of the downstream target of miR-590-5P was performed by the Database for Annotation, Visualization and Integrated Discovery database; D: The binding site of miR-590-5P to METTL3 mRNA 3'untranslated region; E: Western blot assay ensured the analysis on the effect of circ_0001658 and miR-590-5p on the expression of MTL 3; F-G: TTL 3-wild-type and TTL 3-mutant type were co-transfected with mimics or miR-negative control into HT29 and SW480 cells and the luciferase activity was measured; H: The expression of METTL3 mRNA in colorectal cancer (CRC) tissues and paracancerous tissues in 42 cases was probed by quantitative real-time polymerase chain reaction; I-J: The correlation of circ_0001658, miR-590-5p and METTL3 mRNA expression in CRC tissues was analyzed by Pearson's correlation analysis. cP < 0.001. ns: No significance; BP: Biological process; CC: Cellular component; MF: Molecular function; WT: Wild-type; MUT: Mutant type; miR-NC: miRNA negative control; circ-OE: Circ_0001658 plasmid; si-NC: siRNA negative control.
- Citation: Lu Y, Wang XM, Li ZS, Wu AJ, Cheng WX. Hsa_circ_0001658 accelerates the progression of colorectal cancer through miR-590-5p/METTL3 regulatory axis. World J Gastrointest Oncol 2023; 15(1): 76-89
- URL: https://www.wjgnet.com/1948-5204/full/v15/i1/76.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i1.76