Copyright
©The Author(s) 2022.
World J Gastrointest Oncol. Oct 15, 2022; 14(10): 1981-2003
Published online Oct 15, 2022. doi: 10.4251/wjgo.v14.i10.1981
Published online Oct 15, 2022. doi: 10.4251/wjgo.v14.i10.1981
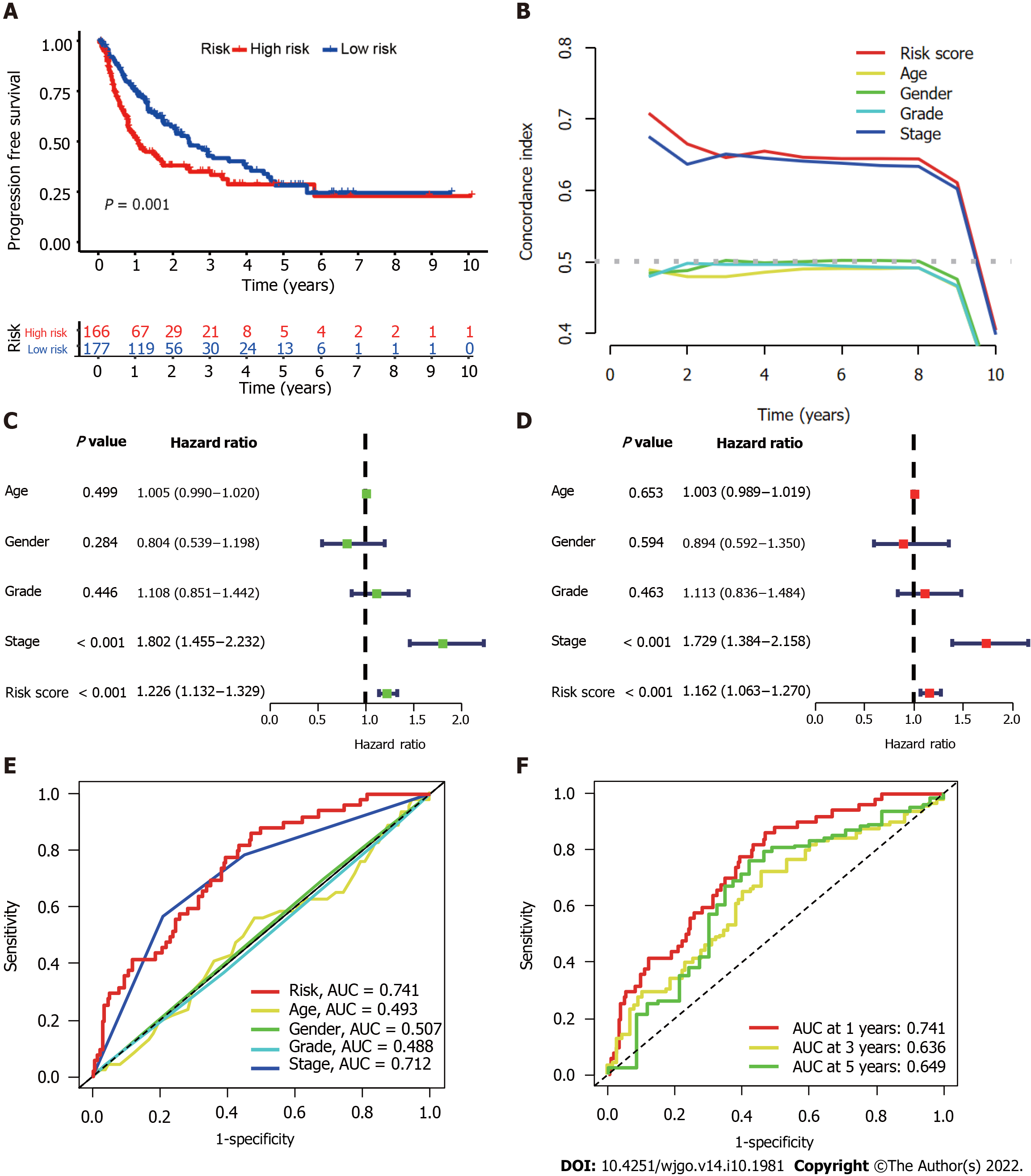
Figure 4 Evaluation of predictive accuracy of the cuproptosis-related long-chain non-coding RNAs signature (CupRLSig) model using the entire The Cancer Genome Atlas-Live Hepatocellular Carcinoma dataset.
A: Kaplan-Meier curves for progression-free survival in high- and low-risk groups stratified by median value of cuproptosis-related long-chain non-coding RNAs signature (CupRLSig) risk score; B: Concordance index curves depicting CupRLSig risk scores and other clinical parameters relevant to predicting hepatocellular carcinoma patient prognosis; C and D: Forest plots for univariate (C) and multivariate (D) Cox proportional-hazards analysis for determination of the independent prognostic value of the CupRLSig risk score; E: Area under the curve (ROC) curves for the CupRLSig risk score and other clinicopathological variables; F: Time-dependent ROC curves for 1-, 3-, and 5-year survival for the CupRLSig signature. ROC: Receiver operating characteristic; AUC: Area under the curve.
- Citation: Huang EM, Ma N, Ma T, Zhou JY, Yang WS, Liu CX, Hou ZH, Chen S, Zong Z, Zeng B, Li YR, Zhou TC. Cuproptosis-related long non-coding RNAs model that effectively predicts prognosis in hepatocellular carcinoma. World J Gastrointest Oncol 2022; 14(10): 1981-2003
- URL: https://www.wjgnet.com/1948-5204/full/v14/i10/1981.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i10.1981