Copyright
©The Author(s) 2021.
World J Gastrointest Oncol. Dec 15, 2021; 13(12): 2129-2148
Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2129
Published online Dec 15, 2021. doi: 10.4251/wjgo.v13.i12.2129
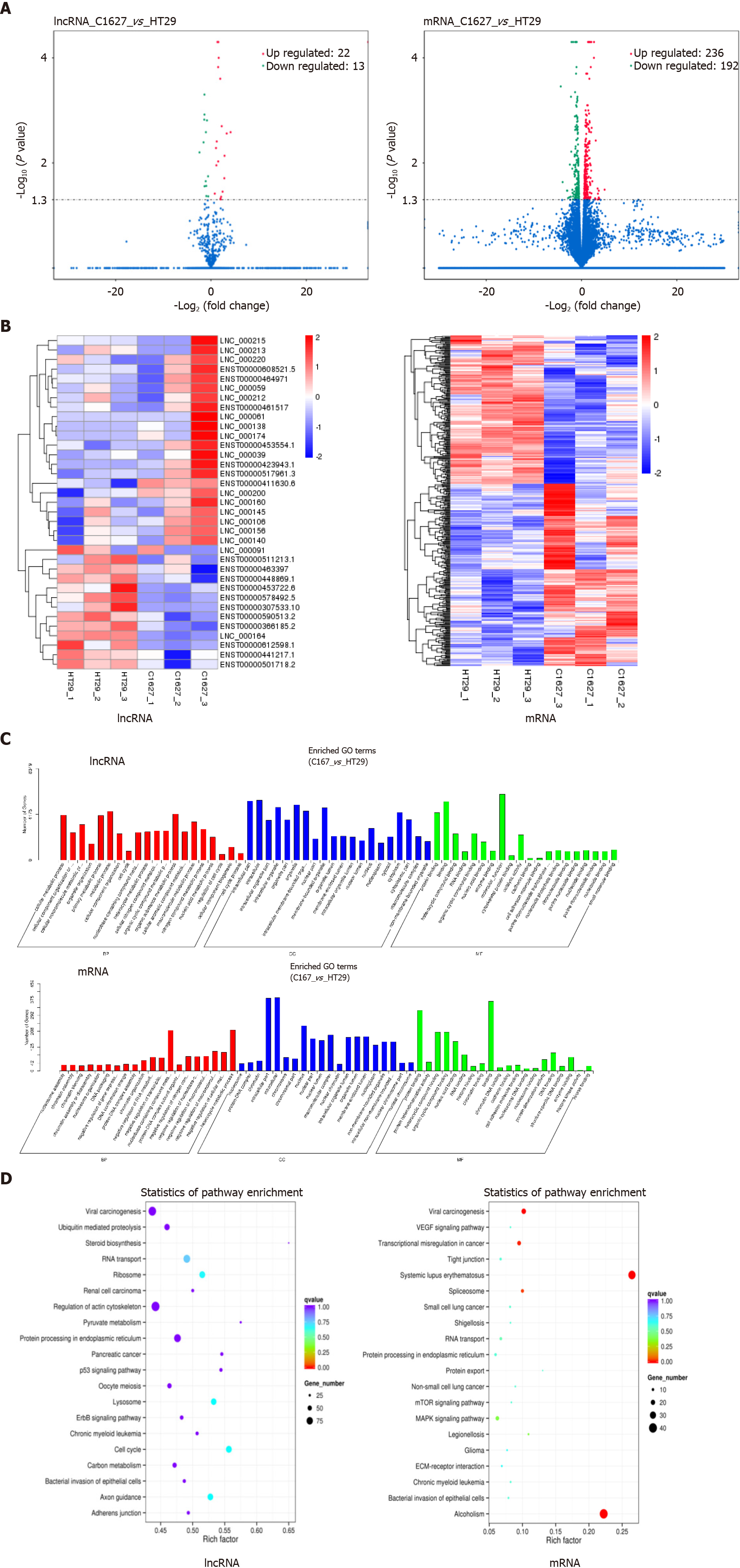
Figure 2 Screening and functional analysis of differentially expressed long noncoding RNAs and mRNAs in exosomes of BRAFV600E mutant colorectal cancer cells and those with BRAF gene silencing.
A: Transcriptomics analysis of differentially expressed long noncoding RNAs (lncRNAs) and mRNAs in the exosomes of HT29 and 1627 cells. Volcano maps are shown (the x axis represents the multiple of difference, and the y axis P values); B: Hierarchical cluster analysis of differentially expressed lncRNAs and mRNAs (red represents upregulated expression, and blue represents downregulated expression); C: GO enrichment analysis of differentially expressed lncRNA target genes and mRNAs; D: KEGG pathway analysis of differentially expressed lncRNA target genes and mRNAs. LncRNA: Long noncoding RNA.
- Citation: Zhi J, Jia XJ, Yan J, Wang HC, Feng B, Xing HY, Jia YT. BRAFV600E mutant colorectal cancer cells mediate local immunosuppressive microenvironment through exosomal long noncoding RNAs. World J Gastrointest Oncol 2021; 13(12): 2129-2148
- URL: https://www.wjgnet.com/1948-5204/full/v13/i12/2129.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i12.2129