Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Apr 15, 2024; 16(4): 1344-1360
Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1344
Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1344
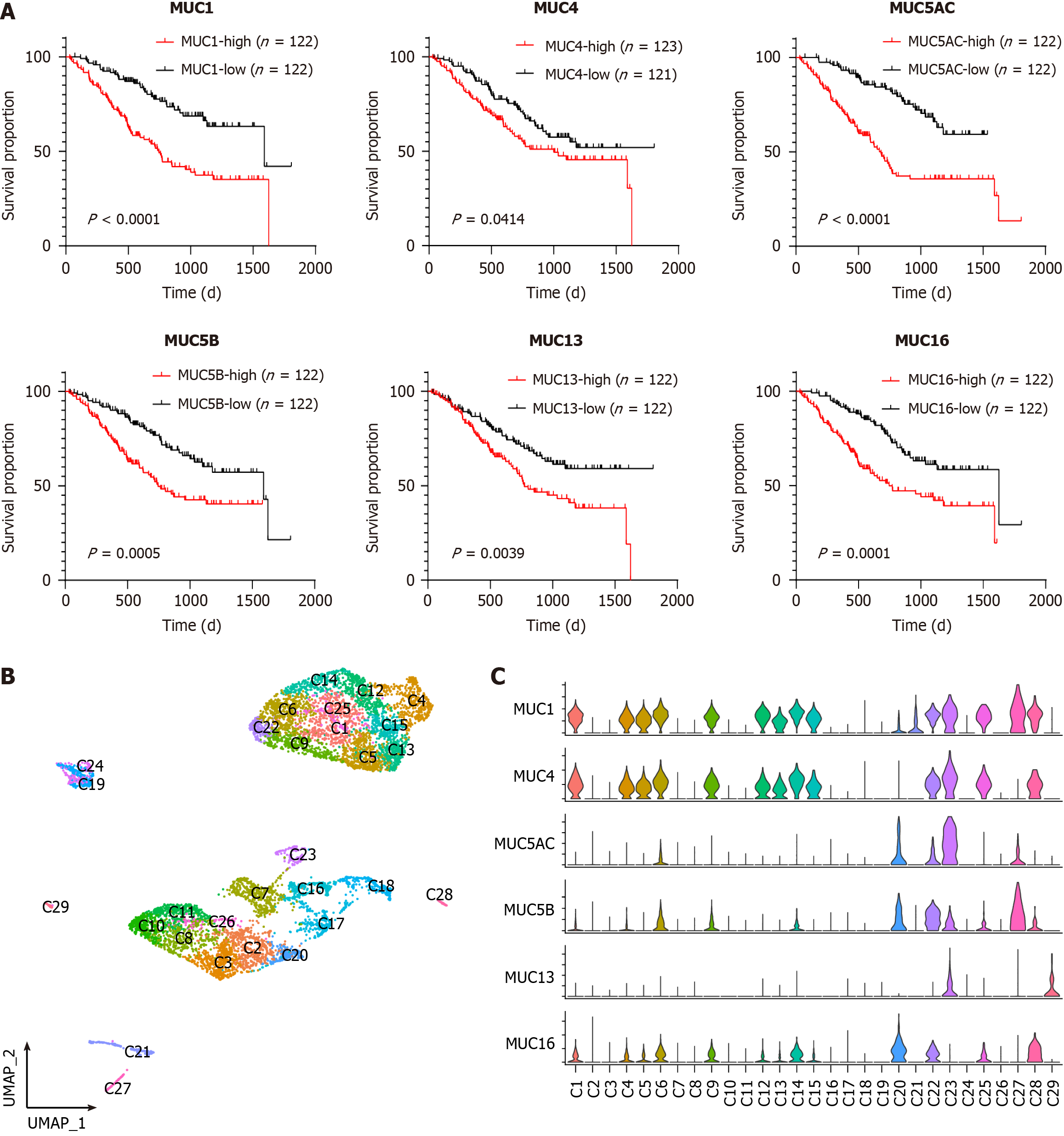
Figure 1 Clinical significance and heterogeneity of mucins in cholangiocarcinoma.
A: Survival curves generated using cohort 2 data illustrating the relationship between cholangiocarcinoma (CCA) patient survival and the mRNA levels of mucins; B: Uniform manifold approximation and projection plot visually representing the tumor cell sub-clusters within the integrated CCA dataset; C: Violin plots depicting of the relative gene expression levels of MUC1, MUC4, MUC5AC, MUC5B, MUC13, and MUC16 in CCA tumor sub-clusters.
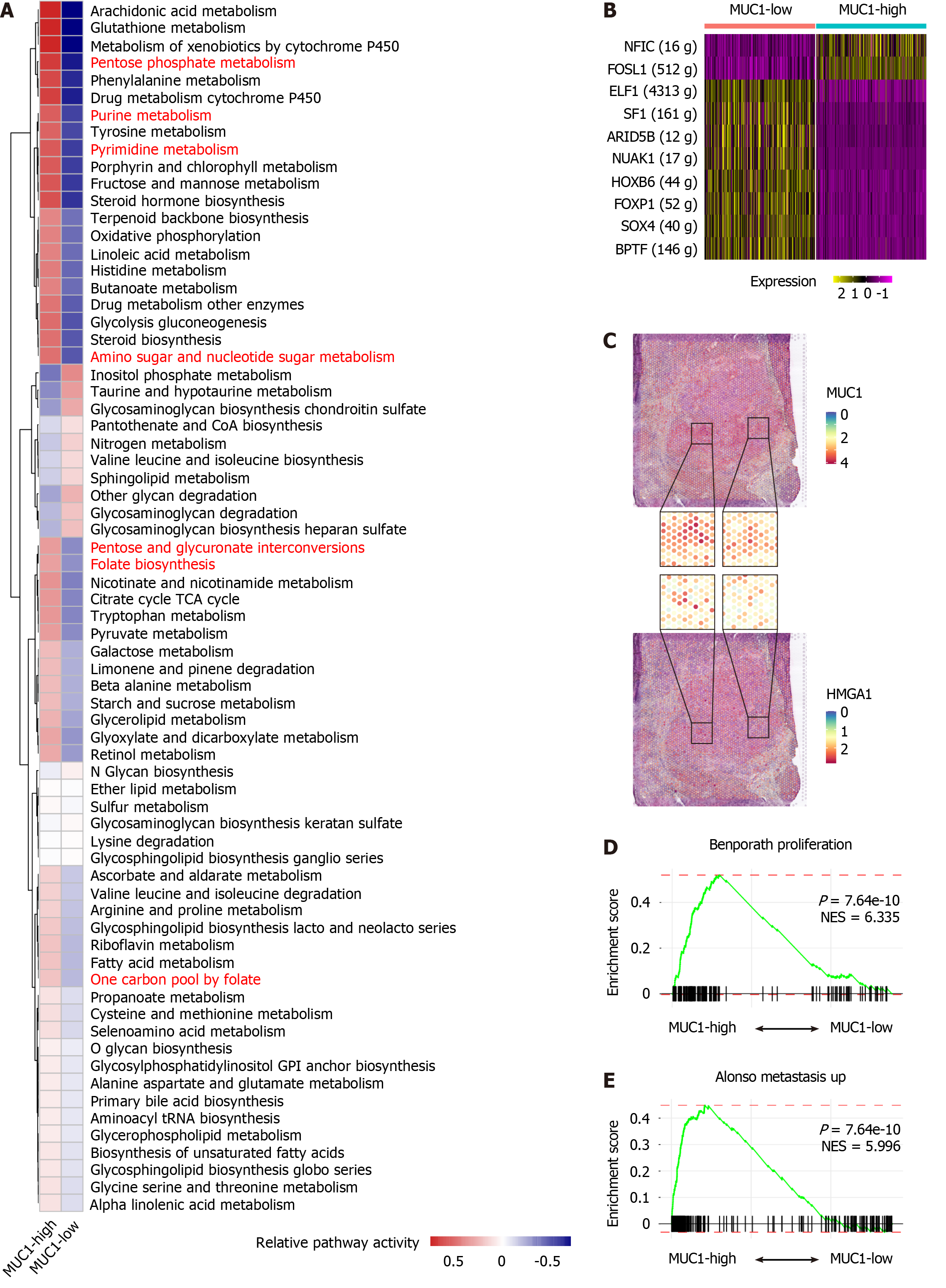
Figure 2 Activated nucleotide metabolic signaling and higher malignant characteristics in MUC1-high tumor cells of cholangiocarcinoma.
A: Heatmap showing the activity of metabolic signaling pathways in MUC1-high vs MUC1-low cells; B: Heatmap showing the different expression levels of transcription factors in MUC1-high cells vs MUC1-low cells; C: Distribution of MUC1 and HMGA1 in the spatial transcriptomics (ST) slide. Zoomed in portions of the ST chip (middle) showing multiple areas of co-localization; D and E: Gene set enrichment analysis plots showing the enrichment of genes from MUC1-high cells (left) vs MUC1-low cells (right) in cell proliferation and metastasis signaling pathways. The P values and normalized enrichment score are indicated on the plots.
Figure 3 WNT signaling and metabolic phenotypes in MUC5AC-high tumor cells of cholangiocarcinoma.
A: Top 40 enriched signaling pathways of the differentially expressed genes in MUC5AC-high cells; B: Relative activity of the WNT signaling pathway among MUC5AC-high, MUC5AC-low, myeloid cells, endothelial cells, fibroblasts, and lymphocytes; C: Violin plots showing relative expression levels of WNT7A, WNT7B, and MMP7 in MUC5AC-high vs MUC5AC-low cholangiocarcinoma (CCA) cells; D: Correlation analysis between the expression of MUC5AC and WNT7B in CCA tumor samples from cohort 10; E: Gene set enrichment analysis plot showing the enrichment of genes from the MUC5AC-high (left) and MUC5AC-low (right) cells in tumor metastasis signaling pathway. The P value and normalized enrichment score are indicated on the plot.
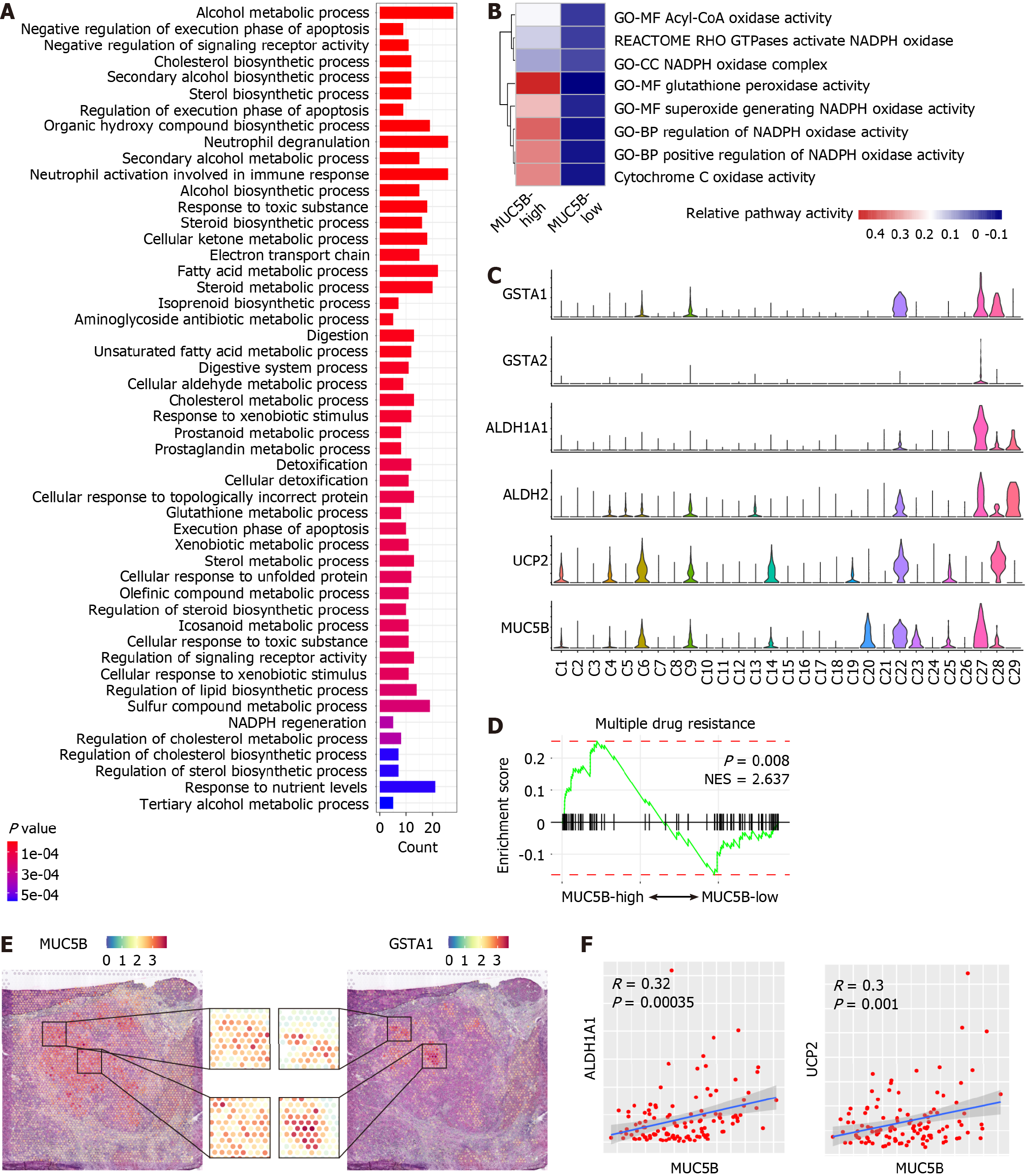
Figure 4 Detoxification function of MUC5B-high tumor cells of cholangiocarcinoma.
A: Top 50 enriched signaling pathways of the differentially expressed genes in MUC5B-high cells; B: Heatmap showing the relative activities of oxidation-related metabolic signaling pathways in MUC5B-high vs MUC5B-low cells; C: Violin plots showing relative expression levels of GSTA1, GSTA2, ALDH1A1, ALDH2, UCP2, and MUC5B in cholangiocarcinoma (CCA) tumor sub-clusters; D: Gene set enrichment analysis plot showing the enrichment of genes from MUC5B-high (left) and MUC5B-low (right) cells in the drug resistance signaling pathway. The P value and normalized enrichment score are indicated on the plot; E: Distribution of MUC5B and GSTA1 in the spatial transcriptomics (ST) slide. Zoomed in portions of the ST chip (middle) showing multiple areas of co-localization; F: Correlation analysis between the expression of MUC5B and ALDH1A1 (left), MUC5B and UCP2 (right) in CCA tumor samples from cohort 10.
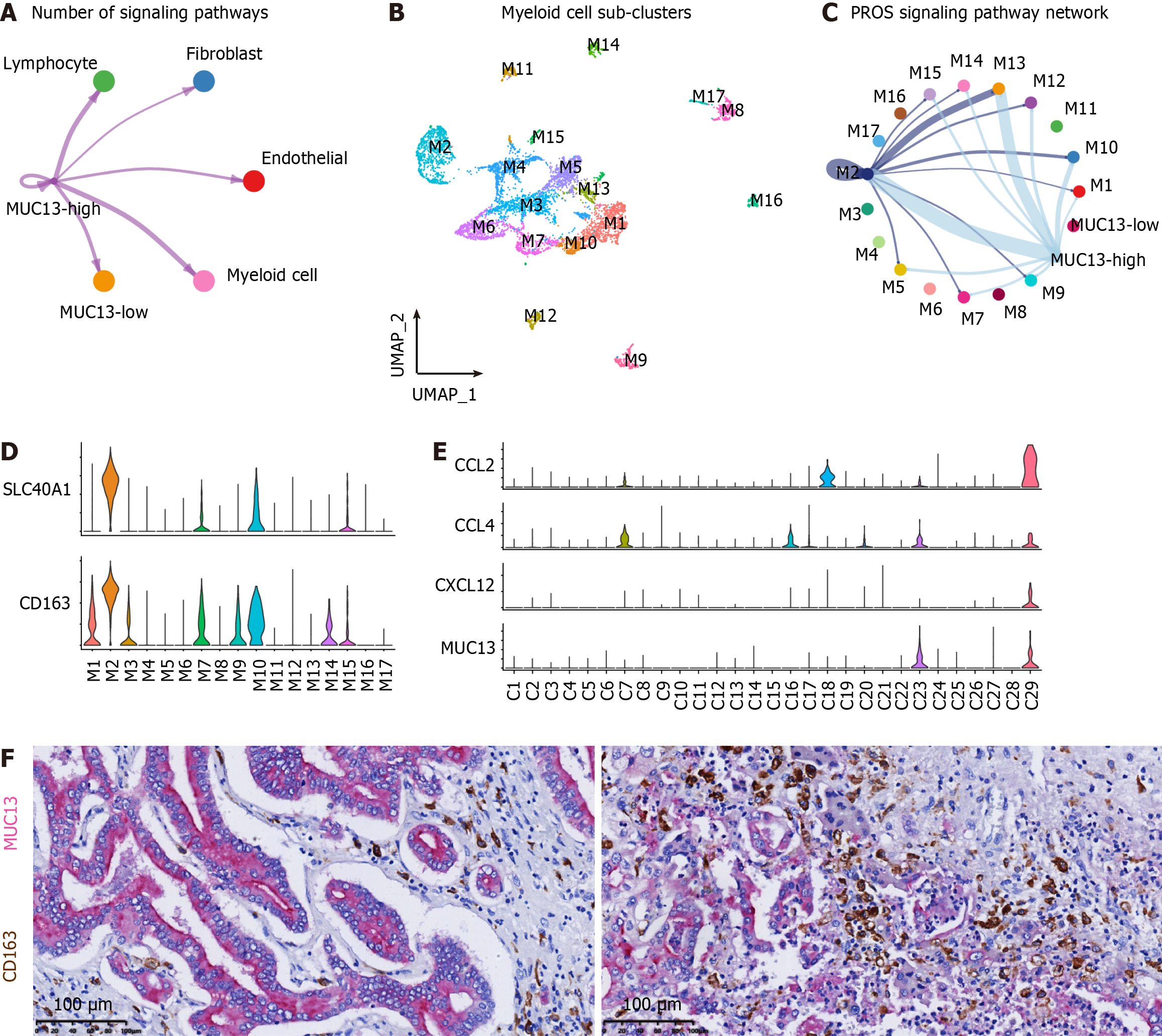
Figure 5 Chemotaxis and M2-polarization of macrophage induced by MUC13-high tumor cells of cholangiocarcinoma.
A: Number of signaling pathways among MUC13-high, MUC13-low, myeloid cells, endothelial cells, fibroblasts, and lymphocytes; B: Uniform manifold approximation and projection plot showing the identification of 17 myeloid cell sub-clusters; C: PROS signaling pathway activity among the MUC13-high sub-cluster, MUC13-low sub-cluster, and myeloid cell sub-clusters; D: Violin plots showing relative expression levels of M2-polarized macrophage marker genes, including SLC40A1 and CD163, in myeloid cell sub-clusters; E: Violin plots showing relative expression levels of CCL2, CCL4, and CXCR12 in tumor cell sub-clusters; F: Immunohistochemistry double staining showing the adjacent distribution of MUC13-high cells (MUC13-positive, purple) and M2-polarized macrophages (CD163-positive, brown) in cholangiocarcinoma. Scale bar, 100 μm.
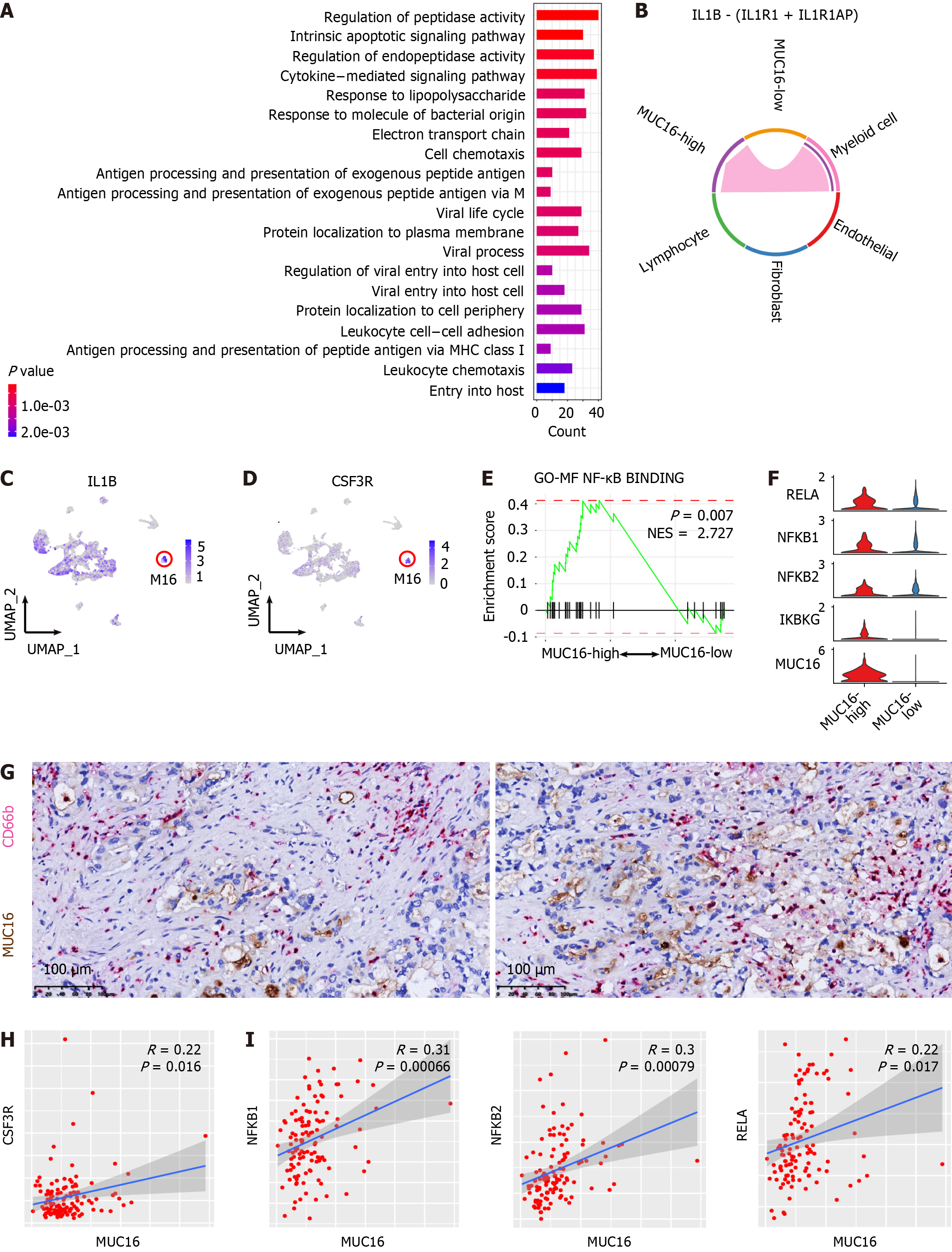
Figure 6 Interaction with neutrophils and activated nuclear factor kappa-light-chain-enhancer of activated B cells signaling in MUC16-high tumor cells of cholangiocarcinoma.
A: Top 20 enriched signaling pathways of the differentially expressed genes of MUC16-high cells; B: Interleukin-1 signaling pathway activity among MUC16-high cells, MUC16-low cells, and cells in tumor microenvironment; C and D: Feature-plots showing the expression level and distribution of IL1B and CSF3R in myeloid cell sub-clusters. Red circles showing the enrichment of IL1B and CSF3R in M16 sub-cluster; E: Gene set enrichment analysis plots showing the enrichment of genes from MUC16-high cells (left) and MUC16-low cells (right) in the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling pathway. The P value and normalized enrichment score are indicated on the plot; F: Violin plots showing relative expression levels of key regulatory genes (RELA, NFKB1, NFKB2, IKBKG) of the NF-κB pathway in MUC16-high vs MUC16-low cells; G: Immunohistochemistry double staining showing the adjacent distribution of MUC16-high cells (MUC16-positive, brown) and neutrophils (CD66b-positive, purple) in cholangiocarcinoma (CCA). Scale bar, 100 μm; H: Correlation analysis between the expression of MUC16 and CSF3R in CCA tumor samples from cohort 10; I: Correlation analysis between the expression of MUC16 and NFKB1, NFKB2, and RELA in CCA tumor samples from cohort 10.
Figure 7 Establishment of cholangiocarcinoma risk evaluation model based on mucin levels.
A: RNA levels of MUC1, MUC13, MUC16, MUC4, MUC5AC and MUC5B in cholangiocarcinoma (CCA) samples from cohort 2; B: Survival curve based on RNA levels of mucins showing the relationship between the risk factor and survival of CCA patients from cohort 2; C-F: Gene set enrichment analysis plots showing the enrichment of differentially expressed genes in high-risk patients in cell proliferation pathway, metastasis pathway, tamoxifen resistance pathway, and cisplatin resistance pathway. The P values and normalized enrichment score are indicated on the plots; G: Survival curve based on protein expression levels of mucins showing the relationship between the risk factor and survival of CCA patients from cohort 2; H-J: Correlation analyses of vascular invasion, regional lymph node metastasis, and tumor node metastasis stage with CCA risk factor; K: Survival curve based on RNA expression levels of mucins showing the relationship between the risk factor and survival of CCA patients from cohort 10.
- Citation: Yang CY, Guo LM, Li Y, Wang GX, Tang XW, Zhang QL, Zhang LF, Luo JY. Establishment of a cholangiocarcinoma risk evaluation model based on mucin expression levels. World J Gastrointest Oncol 2024; 16(4): 1344-1360
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1344.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1344