Copyright
©The Author(s) 2015.
World J Hepatol. Apr 28, 2015; 7(6): 859-873
Published online Apr 28, 2015. doi: 10.4254/wjh.v7.i6.859
Published online Apr 28, 2015. doi: 10.4254/wjh.v7.i6.859
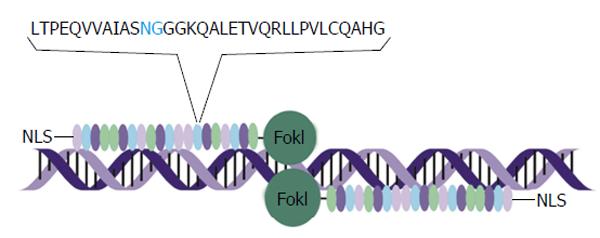
Figure 2 Schematic representation of transcription activator-like effector nuclease interaction with DNA.
A pair of transcription activator-like effector (TALE) nucleases interacts with its target through a tandem arrangement of DNA-binding modules, represented by oval structures in the schematic. Each DNA-binding module comprises 34 amino acids, with a repeat variable diresidue (RVD), in blue, at amino acids 12 and 13. The RVD specifies the DNA base to be targeted. There is a nuclear localisation signal on the N-terminal and a FokI effector bound to the C-terminal. As well as FokI nuclease domains, TALEs may also be bound to transcriptional regulators such as Krüppel-associated box or VP16/64. NLS: Nuclear localisation signal.
- Citation: Nicholson SA, Moyo B, Arbuthnot PB. Progress and prospects of engineered sequence-specific DNA modulating technologies for the management of liver diseases. World J Hepatol 2015; 7(6): 859-873
- URL: https://www.wjgnet.com/1948-5182/full/v7/i6/859.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i6.859