Copyright
©The Author(s) 2015.
World J Hepatol. Feb 27, 2015; 7(2): 150-164
Published online Feb 27, 2015. doi: 10.4254/wjh.v7.i2.150
Published online Feb 27, 2015. doi: 10.4254/wjh.v7.i2.150
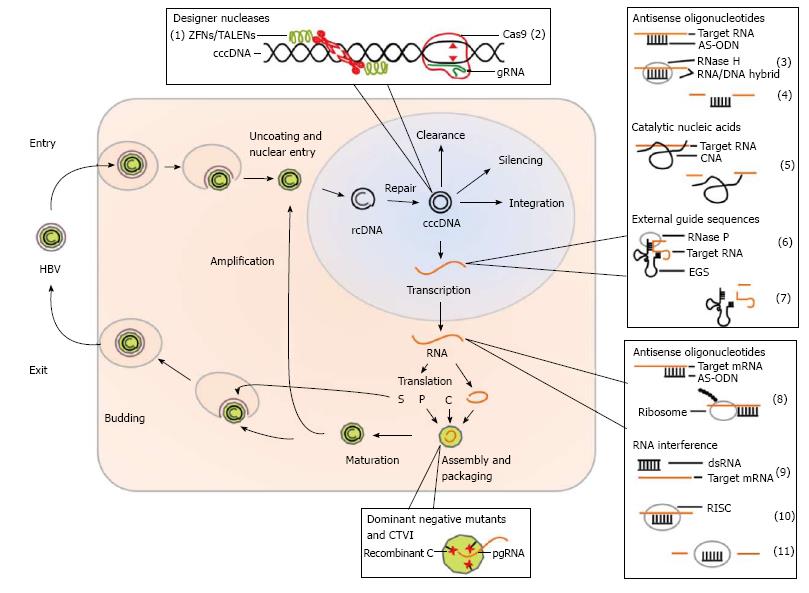
Figure 1 Hepatitis B virus replication cycle and gene therapeutic strategies.
Enveloped virions of hepatitis B virus (HBV) infect liver cells via attachment to the cell membrane and endocytosis. The capsid with the relaxed circular (rc) DNA is released into the cytoplasm and the DNA is uncoated upon nuclear entry. In the nucleus the rcDNA is repaired to form the covalently closed circular (ccc) DNA. The cccDNA can be eventually cleared out, silenced or integrated into the host genome. Predominantly, it persists as an episome in the nucleus and is transcribed and translated by the host cell machinery. One of the transcripts forms the pregenomic (pg) RNA which is encapsulated together with the translated viral polymerase (P) by the translated viral capsid proteins (C). In the newly assembled nucleocapsid the pgRNA serves as a template for the viral polymerase which synthesizes the rcDNA. The nucleocapsid either migrates back to the nucleus to increase the pool of cccDNA or is internalized by the endoplasmatic reticulum (ER). In the latter process it is enveloped with ER-membrane that harbors translated viral surface proteins (S) and finally released from the cell. Gene therapeutic strategies act on several steps of the viral replication cycle. Designer nucleases are intended to promote disruption of the cccDNA. Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) (1) use protein-based DNA-binding modules, while the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 nuclease system (2) is directed by a guide RNA (gRNA) to the target site. Antisense oligonucleotides (AS-ODNs) act inhibitory on the viral replication in two ways. In the nucleus they recruit RNase H (3) after the formation of RNA/DNA hybrids, which cleaves the viral RNA (4). In the cytosol they bind to the viral RNA which leads to a steric blockade of subsequent processes (8). Catalytic nucleotides (CNA) as ribozymes and DNAzymes are able to cleave targeted RNA by themselves (5). External guide sequences (EGS) are designed in a way that they resemble precursor tRNAs (6) when they bind to their target RNA and trigger cleavage by RNase P (7). RNA interference can be induced by different dsRNA species (9). Longer dsRNAs can be delivered exogenously or expressed in the cells as shRNAs. The RNAs are processed to approximately 21 nucleotide long dsRNAs termed siRNAs which are used to form the large RNA-induced silencing complex (RISC) (10) which degrades target messenger RNAs (mRNAs) (11). Dominant negative mutants of capsid proteins form a hindrance for proper packaging of viral progenitor RNA. In contrast, capsid-targeted viral inactivation (CTVI) is an approach where the capsid proteins are additionally fused to a destructive compound.
- Citation: Gebbing M, Bergmann T, Schulz E, Ehrhardt A. Gene therapeutic approaches to inhibit hepatitis B virus replication. World J Hepatol 2015; 7(2): 150-164
- URL: https://www.wjgnet.com/1948-5182/full/v7/i2/150.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i2.150