Copyright
©2014 Baishideng Publishing Group Inc.
World J Hepatol. Dec 27, 2014; 6(12): 870-879
Published online Dec 27, 2014. doi: 10.4254/wjh.v6.i12.870
Published online Dec 27, 2014. doi: 10.4254/wjh.v6.i12.870
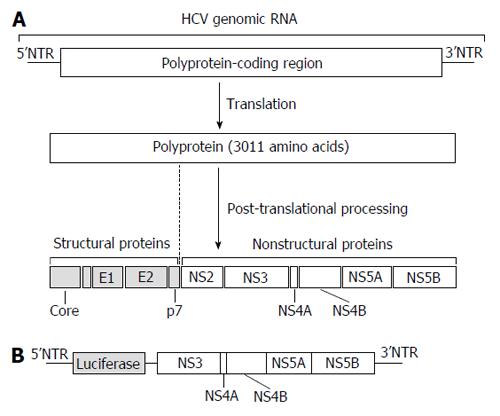
Figure 1 Structure of the hepatitis C virus genome and cell system for anti-hepatitis C virus drug discovery.
A: HCV genomic RNA and viral proteins. HCV genomic RNA encodes a single polyprotein of 3011 amino acids. After being translated, the polyprotein is processed into 4 structural proteins (Core, E1, E2, and p7) and 6 non-structural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The polyprotein-coding region is flanked by 5’ and 3’NTRs. Viral RNA also serves as a template for viral genome replication and both NTRs modulate viral protein synthesis and genome replication; B: The HCV replicon cell system. Huh-7 cells were transfected with the luciferase gene connected with HCV subgenomic RNA including the downstream coding regions of NS3. The expression of HCV subgenomic RNA could be quantified by luciferase activity. HCV: Hepatitis C virus; NTRs: Non-translated regions.
- Citation: Ishida YI, Takeshita M, Kataoka H. Functional foods effective for hepatitis C: Identification of oligomeric proanthocyanidin and its action mechanism. World J Hepatol 2014; 6(12): 870-879
- URL: https://www.wjgnet.com/1948-5182/full/v6/i12/870.htm
- DOI: https://dx.doi.org/10.4254/wjh.v6.i12.870