Copyright
©The Author(s) 2025.
World J Hepatol. Feb 27, 2025; 17(2): 99046
Published online Feb 27, 2025. doi: 10.4254/wjh.v17.i2.99046
Published online Feb 27, 2025. doi: 10.4254/wjh.v17.i2.99046
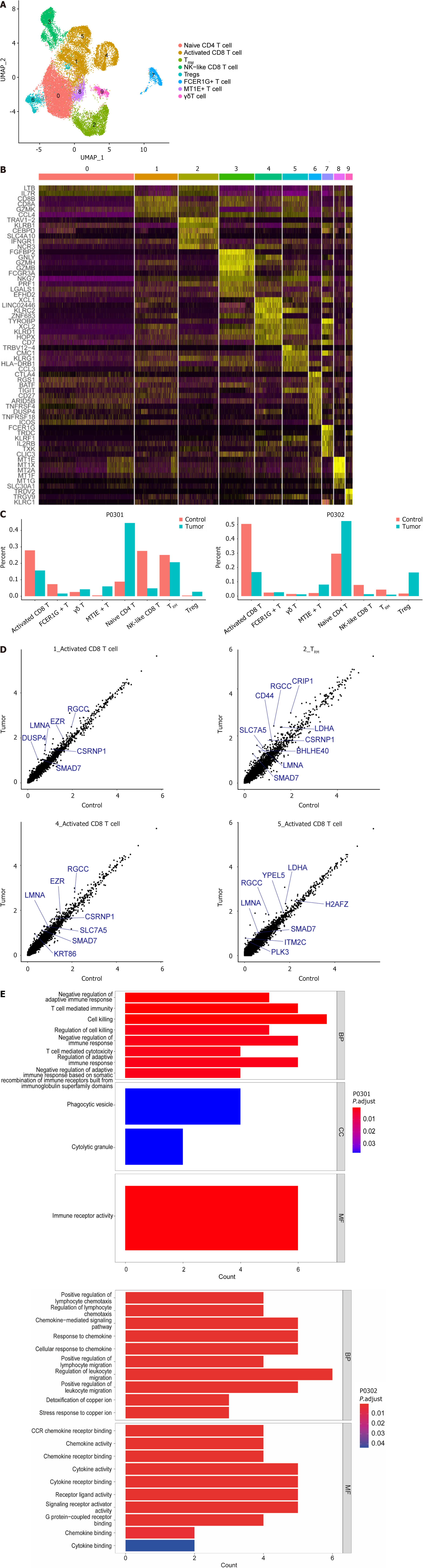
Figure 2 Comprehensive analysis of cancer and paracancerous T cells in hepatocellular carcinoma patients.
A: Cluster distribution of T-cell subsets on uniform manifold approximation and projections after subdivision; B: Marker gene heatmap of the subpopulations. The horizontal line represents the marker gene name, with the cells listed vertically and the upper color bar representing different subpopulations; C: Histogram showing the difference in the proportions of T-cell subsets between cancer patients and paracancerous patients; D: Scatter plot of differential expressed genes showing genes highly expressed in tumor 1_Activated CD8+ T cells, 2_TRM, 4_Activated CD8+ T cells and 5_Activated CD8+ T cells at P0301. Each dot represents a gene; E: Gene ontology (GO) enrichment analysis results showing the functional enrichment of upregulated differential expressed genes s in tumor tissues. The vertical axis is the GO entry, and the horizontal axis is the number of genes enriched in the GO entry. The redder color indicates greater significance. UMAP: Uniform manifold approximation and projections; TRM: Tissue-resident memory T cells; NK: Natural killer; Treg: Regulatory T cells; FCER1G: Fc epsilon receptor 1G; MT1E: Metallothionein 1E; BP: Biological process; CC: Cellular composition; MF: Molecular function.
- Citation: Gu XY, Gu SL, Chen ZY, Tong JL, Li XY, Dong H, Zhang CY, Qian WX, Ma XC, Yi CH, Yi YX. Uncovering immune cell heterogeneity in hepatocellular carcinoma by combining single-cell RNA sequencing with T-cell receptor sequencing. World J Hepatol 2025; 17(2): 99046
- URL: https://www.wjgnet.com/1948-5182/full/v17/i2/99046.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i2.99046