Copyright
©The Author(s) 2024.
World J Hepatol. Aug 27, 2024; 16(8): 1145-1155
Published online Aug 27, 2024. doi: 10.4254/wjh.v16.i8.1145
Published online Aug 27, 2024. doi: 10.4254/wjh.v16.i8.1145
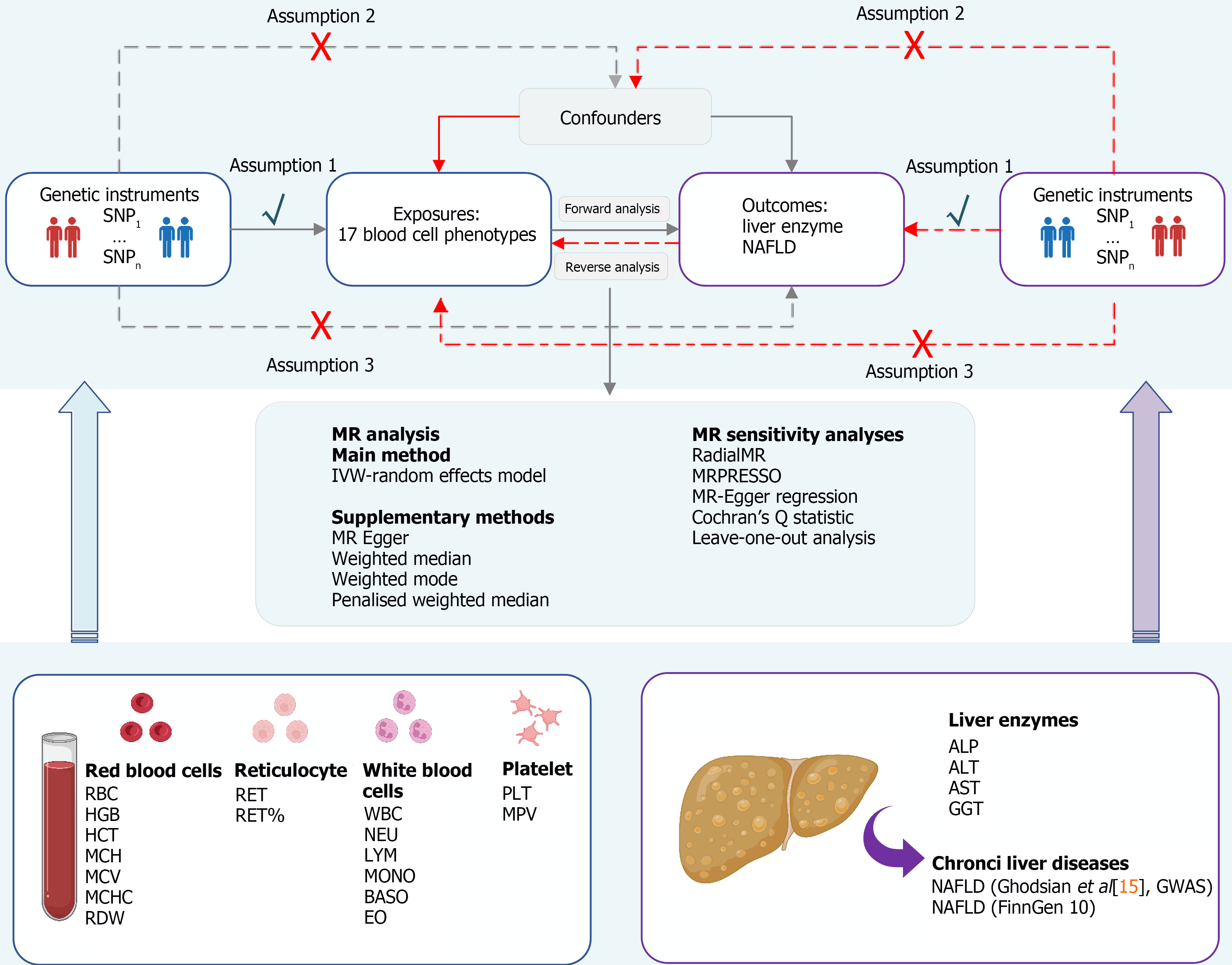
Figure 1 Mendelian randomization assumptions and study design.
Mendelian randomization (MR) analysis relies on three key assumptions. Assumption 1: Genetic variants are closely associated with exposure (blood cell traits); Assumption 2: Genetic variants are not associated with any potential confounders; Assumption 3: Genetic variants are not associated with outcome (liver enzymes and nonalcoholic fatty liver disease) except via exposure. MR: Mendelian randomization; MRPRESSO: Mendelian randomization pleiotropy residual sum and outlier; RBC: Red blood cell count; HGB: Haemoglobin concentration; HCT: Haematocrit; MCH: Mean corpuscular haemoglobin; MCV: Mean corpuscular volume; MCHC: Mean corpuscular haemoglobin concentration; RDW: Red cell distribution width; WBC: White blood cell count; NEU: Neutrophil count; LYM: Lymphocyte count; MONO: Monocyte count; BASO: Basophil count; EO: Eosinophil count; PLT: Platelet count; MPV: Mean platelet volume; RET: Reticulocyte count; RET%: Reticulocyte fraction of red cells; ALP: Alkaline phosphatase; ALT: Alanine aminotransferase; GGT: Gamma-glutamyl transferase; AST: Aspartate aminotransferase; NAFLD: Nonalcoholic fatty liver disease. Created with BioRender.com.
- Citation: Hu B, Wan AH, Xiang XQ, Wei YH, Chen Y, Tang Z, Xu CD, Zheng ZW, Yang SL, Zhao K. Blood cell counts and nonalcoholic fatty liver disease: Evidence from Mendelian randomization analysis. World J Hepatol 2024; 16(8): 1145-1155
- URL: https://www.wjgnet.com/1948-5182/full/v16/i8/1145.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i8.1145