Copyright
©The Author(s) 2024.
World J Hepatol. Jun 27, 2024; 16(6): 932-950
Published online Jun 27, 2024. doi: 10.4254/wjh.v16.i6.932
Published online Jun 27, 2024. doi: 10.4254/wjh.v16.i6.932
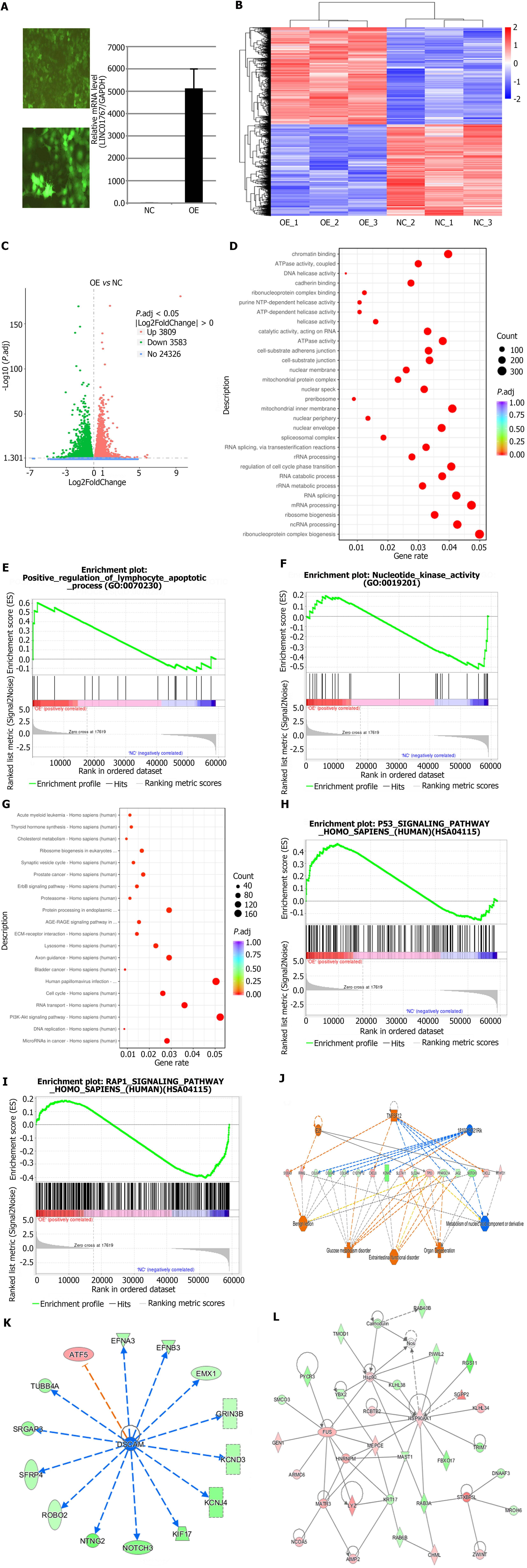
Figure 3 RNA-sequencing and bioinformatic analysis in Huh7 with LINC01767 over expression.
A: Over expression of LINC01767 in Huh7 cells; B: The differential expression heatmap is presented in using the log fold change (FC) > 1.5, and P < 0.05; C: The differential expression volcano plot is presented in using the logFC > 1.5, and P < 0.05; D: The top 10 most significantly enriched Gene Ontology terms; E and F: Gene Set Enrichment Analysis (GSEA) analysis showed that go term of LINC01767 involved in positive regulation of lymphocyte apoptosis, nucleotide kinase activity and so on; G: The top 10 most significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) terms; H and I: GSEA analysis showed KEGG pathway showed LINC01767 was involved in P53 signaling, RAP1 signaling and other important pathways; J: The top-ranked regulatory network in this regulatory effect analysis shows that the regulation net involved in regulators 1810019D21Rik, E2f, TNFSF12 through C1QTNF12, COL6A1, COL6A2, COL9A3, CXCL2, CXCL8, JAG2, KCNH2, MTHFD1, NOTCH3, PPARGC1A, RRM2, S100A9, SLC2A4, SLC7A11, TP53; K: The updream regulatory network analysis shows the DSCAM and ATF5 and so on involved in the regulatory net; L: Molecular network showed it can interact with HSP90, LYZ, FUS, and so on.
- Citation: Zhang L, Cui TX, Li XZ, Liu C, Wang WQ. Diagnostic and prognostic role of LINC01767 in hepatocellular carcinoma. World J Hepatol 2024; 16(6): 932-950
- URL: https://www.wjgnet.com/1948-5182/full/v16/i6/932.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i6.932