Copyright
©The Author(s) 2024.
World J Hepatol. Dec 27, 2024; 16(12): 1468-1479
Published online Dec 27, 2024. doi: 10.4254/wjh.v16.i12.1468
Published online Dec 27, 2024. doi: 10.4254/wjh.v16.i12.1468
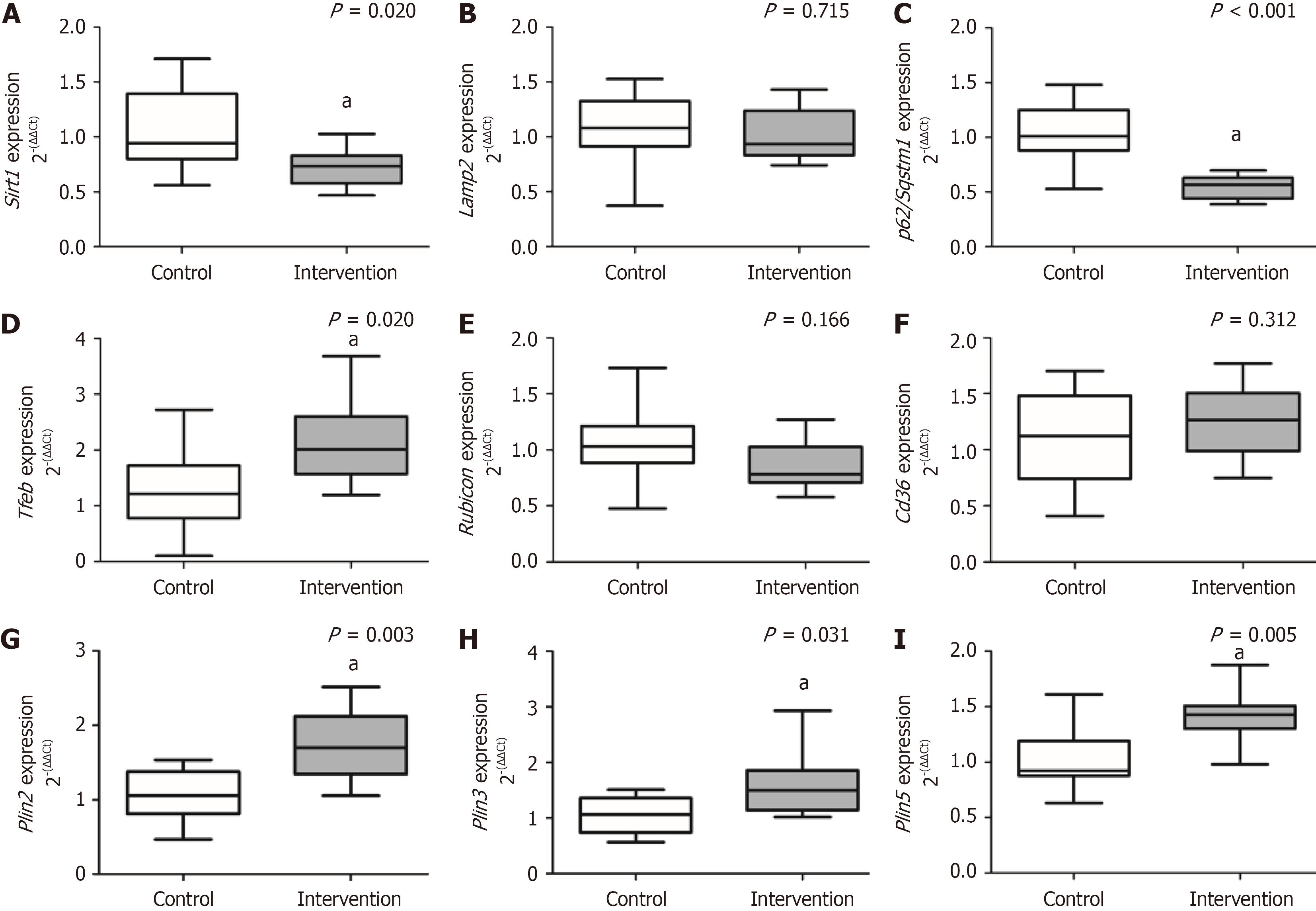
Figure 3 Hepatic gene expression of lipophagy markers.
A: Sirtuin-1; B: Lysosome-associated membrane protein-2; C: P62/sequestosome-1; D: Transcription factor EB; E: Rubicon; F: Cd36; G: Perilipin 2 (Plin2); H: Plin3; I: Plin5. aP < 0.05 considered significant, Mann-Whitney U test. Variables are described as median (25th-75th percentiles). Lamp2: Lysosome-associated membrane protein-2; Plin: Perilipin; p62/Sqstm1: P62/sequestosome-1; Sirt1: Sirtuin-1; Tfeb: Transcription factor EB.
- Citation: Schütz F, Longo L, Keingeski MB, Filippi-Chiela E, Uribe-Cruz C, Álvares-da-Silva MR. Lipophagy and epigenetic alterations are related to metabolic dysfunction-associated steatotic liver disease progression in an experimental model. World J Hepatol 2024; 16(12): 1468-1479
- URL: https://www.wjgnet.com/1948-5182/full/v16/i12/1468.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i12.1468