INTRODUCTION
In mammals, liver is the only organ that possesses the strong capability of regeneration[1-7]. Since Higgins and Anderson[8] established the model of partial hepatectomy (PH), it has been widely used to study the reconstruction and regeneration of tissue and organs, cellular multiplication, dedifferentiation, stress response and the regulation of physiology and biochemistry[8-16]. But due to the complexity of liver regeneration, the molecular mechanism of LR remains to be elucidated. In order to better understand the mechanism of PH, researchers have established a series of successive partial hepatectomy (SPH) models. These included the long interval successive partial hepatectomy (LISPH) that involved an interval of more than three weeks and short interval successive partial hepatectomy (SISPH) which had an interval of 4 and/or 36 hr[17]. Because the hepatocytic initiation, dedifferentiation, division and redifferentiation mainly occured within 144 hr following PH[18-22]. It is doubtless that the study on the change of liver tissue, cellular morphology, structure, physiology and biochemistry in every crucial point following SISPH can offer more data about LR. In this paper, we used the normal liver tissue as control and the liver tissue in 112 hr SISPH as experimental materials, and adopted the whole-mount research strategy of genome and resorted to SSH technique to construct a differential expressed forward-subtracted cDNA library. From the library, we picked some significant expressed sequence tags (ESTs) clones. Then the primers were designed according to these ESTs. By RACE PCR, some full-length cDNAs were obtained. After sequencing these full-length cDNAs, it showed that one of them belonged to the gene of related transthyretin, termed LR1 gene, which was up-regulated expression in LR.
MATERIALS AND METHODS
Making SISPH model and preparation of samples
Adult Spargue-Dawfey rats (weighing 200-250 g) were provided by the experimental animal house of Henan Normal University and 0-4-36-36-36 hr SISPH model was made according to the method of Xu et al[17]. Lobus external sinister and lobus centralis sinister, lobus centralis, lobus dexter, lobus candatus were removed one by one at four different time points 4, 36, 36 and 36 hr (total time: 4 hr, 40 hr, 76 hr, 112 hr) respectively. The control and the four resected liver hepatic lobus were washed with precooled phosphate-buffered saline (PBS) thoroughly before these samples were frozen in liquid nitrogen and transferred to the -80 °C freezer for storage.
Preparation of control mRNA and the 112 hr SISPH mRNA
Isolation of total RNA and mRNA was carried out according to the protocol of RNAease kit and oligotex mRNA spin column purification kit (Qiagen). The quantity and integrity of mRNA were detected by ultraviolet spectrometer and by electrophoresing on a denaturing formaldehyde agarose stained by EtBr. Chemicals and reagents used were at least of analytical grade.
Generating of a subtracted cDNA library
SSH was performed between the aforementioned tester and driver liver tissue mRNA preparations using a PCR selectTM cDNA subtraction kit and 50 × PCR enzyme kit (Clontech, Heidelberg, Germany) following the instructions of the manufacturer. Briefly, 2 µg aliquots each of poly (A+) mRNA from the tester and the pooled driver were subjected to cDNA synthesis. Thereafter, the tester and driver cDNAs were digested with Rsa I. The tester cDNA was subdivided into two portions, and each was ligated with a different cDNA adaptor. In the first hybridization reaction, an excess of driver was added to each sample of tester. The samples were heat denaturated and allowed to anneal. Due to the second-order kinetics of hybridization, the concentration of high- and low-abundance sequence is equalized among the single-stranded tester molecules. At the same time, single-stranded tester molecules are significantly enriched for differentially expressed sequences. During the second hybridization, the two primary hybridization samples were mixed together without denaturation, only the remaining equalized and subtracted single-stranded tester cDNAs can reassociate to form double-stranded tester molecules with different ends. After filling in the ends with DNA polymerase, the entire population of molecules was subjected to nested PCR with two adaptor-specific primer pairs. The tester control was amplified with primer1 and nested primer 1 and 2 provided in the kit. The subtraction efficiency was evaluated by house-keeping gene g3pdh.
Cloning the subtracted library into a TA vector
Products from the secondary PCR reactions were ligated into a pGEM-T vector (Promega) and the resultant ligation products were then transformed into JM109 E. coli competent cells, The bacterial were subsequently grown in 800 ul of liquid Luria-Bertani medium and allowed to incubate for 45 min at 37 °C with shaking at 150 rpm. Thereafter, the cells were plated onto agar plates containing ampicillin (50 μg/ml), 5-bromo-4-chloro-3-indoly-β-D-galactoside (x-gal; 20 μg/cm2) and isoploprl-β-D-thiogalactoside (IPTG; 12.1 μg/cm2) and incubated overnight at 37 °C. Individual recombinant white clones were picked and grown in single line pattern onto Luria-Bertani agar solid medium containing ampicillin and allowed to incubate at 37 °C for 6-7 hr before single clone was picked from single-line pattern agar medium and allowed to grow in Luria-Bertani liquid medium containing ampicillin overnight at 37 °C with shaking of 150 rpm.
Identifying the subtracted clones
Plasmids were isolated from bacterial clones harboring differentially expressed cDNA sequences as described by Sambrook et al Plasmids containing cDNA inserts were amplified by PCR with nested primer 1 and primer 2 and the amplified product of every plasmid was detected by agarose gel electrophoresis. After that, the inserts that appeared as distinct bands were sequenced by T7/SP6 chain termination reation in TaKaRa (DaLian, China). Nucleic acid homology searches were subsequently performed at the National Center of Biotechnology Information (National Institutes of Health, Bethesda, Md., NCBI).
RACE
An interested EST fragment was chosen from the established cDNA library for primer design according to the instructions of the 5’/3’ RACE kit (Roche) manufacturer. RACE, a procedure to amplify nucleic acid sequences from a mRNA template between a defined internal site and unknown sequences at 5’-and 3’-end of mRNA, was then performed by using 5’/3’ RACE kit and following the supplier’s protocol. The primer used for 3’ RACE was SP5: 5’-AGCTGGGAGCCGTTTGC-3’ and the primer used for 5’ RACE was SP1: 5’-TGCTGTAGGAGTACGGGCTC3’, SP2: 5’-ACCAGAGTCATTGGCTGT-3’, SP3: 5’-GCCCGTGCAGCTCT-3’ respectively. Proofreading pfu DNA polymerase was purchased from MBI and the 3’ RACE PCR parameters were: predenaturation at 94 °C for 2 min before amplification was done for 40 cycles at 94 °C for 45 sec, annealing at 65 °C for 1 min and extension at 72 °C for 1 min and 30 sec followed by an additional extension period at 72 °C for 5 min. The 5’ RACE used touchdown PCR and amplification was carried out with the following conditions: 94 °C for 2 min, 25 cycles including a denaturation step at 94 °C for 15 sec, an annealing step from 68 °C to 55 °C for 30 sec, and extension at 72 °C for 1 min and 30 sec; 15 cycles as above except that the annealing step at 55 °C; a final extension step at 72 °C for 7 min ended the PCR reaction. The products were subjected to electrophoresis in a 1% agarose gel, purified according to qiaquick PCR purification kit protocol and added dATP-tailing to blunt-ended fragments at 70 °C for 30 min in the presence of Taq DNA polymerase before ligated into the pGEM-T vector. Transformation, screening, and sequencing were then performed as described above.
Splicing the full-length nucleotide sequence
Alignment of the sequences was spliced, in light of contig of 5’/3’ sequencing result, to get a full-length nucleotide sequence. Homology search was performed using the BLAST program at NCBI.
RESULTS
Construction of the subtracted cDNA library and screening of the differential fragments
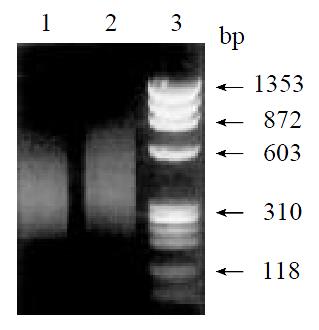
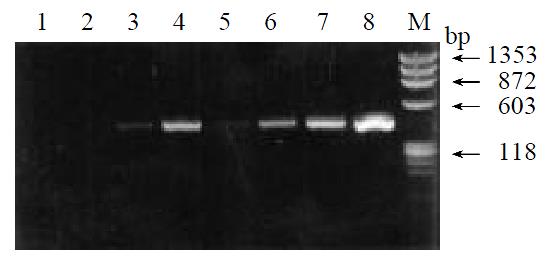
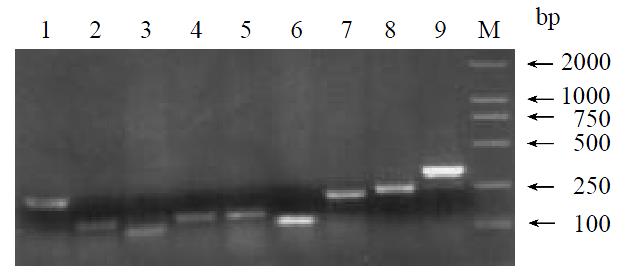
By means of suppression subtractive hybridization (SSH) technique, we prepared driver cDNAs and tester cDNAs of liver tissue from control and liver tissue 112 hr following SISPH. After two rounds hybridization and two PCR reactions, we obtained the forward-subtractive cDNA library (Figure 1). The subtracted cDNA was evaluated by the house-keeping gene g3pdh (Figure 2). Our results showed that the constructed forward-subtracted cDNA library was highly efficient after cDNA library was subcloned into pGEM-T vector and transformed into JM109, we obtained 120 white clones and 40 clones randomly selected were then subjected to sequencing (Figure 3). The sequencing results were sent to NCBI for BLAST searches.
Figure 1 Gel electrophoresis of SSH results.
1. Control; 2. Forward subtractive; 3. Marker.
Figure 2 Effiency of subtracted library.
1-4. Subtracted products with g3pdh primer; 5-8. Control products with g3pdh primer; M. Marker.
Figure 3 Gel electrophoresis of clones inserted fragments.
1-9. Different EST fragments; M. Marker.
Determination of the full-length nucleotide sequence of LR1 cDNA by RACE
The original cDNA isolated from the cDNA subtraction screening was about 311 bp long. This cDNA fragment was used to design primers according to 5’/3’ RACE kit. After amplification, ligation, screening and sequencing, we obtained the sequence of the 3’ end and 5’ end (Figure 4). Alignment of contig sequences enabled us to delineate the entire cDNA sequence (Figure 5). We found a single long ORF starting with a methionine codon ATG at nucleotide 47 and ending with a TAA stop codon at 625 and a putative polyadenylation signal AATAAA upstream from the poly (A+). Homology search using LR1 cDNA sequence revealed that it is 88.8% homologous to rat transthyretin gene and 88.4% homologous to albumin gene. Protein homology (Swissport database) search of the peptide encoded by the LR1 ORF revealed that it is 91% homologous to rat transthyretin, submitting this full-length cDNA to NCBI and obtain GenBank Accession No. AF479660.
Figure 4 RACE PCR results.
1.3’ RACE result; 2.5’ RACE result; 3. Marker.
Figure 5 Full-length cDNA of LRI
DISCUSSION
SSH technique was developed based primarily on a recently described technique called cDNA suppression PCR which combines normalization and subtraction in a single procedure to generate subtracted cDNA library[23-25]. Compared to the methods of mRNA differential display[26], representational difference analysis of DNA[27], it has two distinct advantages: (1) it has a high subtraction efficiency; (2) it harbors an equalized representation of differentially expressed sequences which can separate effectively both high and low copy expressed genes mainly because of normalization. The normalization step equalizes the abundance of cDNAs within the target population and the subtraction step excludes the common sequences between the target and the driver populations[23,28,29]. The experimental results showed the SSH technique can enrich for rare sequences over 1000-5000 fold in one round of subtractive hybridization[23,24,30]. By resorting to SSH and high-density array scanning technique, using non-metastatic counterpart Bsp73-1AS as driver, metastatic adenocarcinoma cell line Bsp73-ASML as tester, Von Stein[31] found up to 252 over-expressed clones among 625 clones with no homologous sequences in GenBank. By using a EST fragment derived from the forward-subtracted cDNA library as probe, we have cloned a novel gene from the liver tissue 112 hr cDNA library following SISPH. These results suggest that SSH technique is applicable to many molecular genetic and positional cloning studies for the identification of disease, developmental, tissue-specific or other differentially expressed genes.
With the aim to shed light on the molecular mechanism of PH, Xu et al[17] established the SISPH model which selected the intervals at the crucial periods, e.g. the peak of cell activation (4 hr after PH), the peak of cell division (36 hr after PH) and the peak of re-differention (48 hr after PH). We have examined some biological activated factors, heat shock proteins (HSPs), proteases, proteins related to cell proliferation and dephosphoralization[13,14,17,18,32]. These data have provided useful information to reveal the molecular mechanism of LR. The results suggested that SISPH model may offer more abundant data than that of PH. In present study, We took 0 h and 112 hr as driver and tester respectively in 0-4-36-36-36hr SISPH model when resorting to SSH strategy, then we constructed a forward-subtractive cDNA library in short time by using RACE, subsequently we cloned a full-length cDNA and blast search showed that the cDNA is 88.8% and 88.4% homologous to the transthyretin and albumin in rat respectively. This suggested that the gene may exert partial function idendifical to transthyretin. It is well known that thyroxin and its metabolic products integrated thyroxine-binding prealbumin and albumin is derived from liver to operate. In the physiological conditions, 99.9% 3,3',5,5'-tetraiodothyronine (T4) and 99.7% 3,3'5-triiodothyronine (T3) are complexed with proteins which contain over 60% tranthyretin and 15%-30% thyroxine-binding prealbumin and 10% albumin. Our present study showed that the injured liver released abundant analogs of transthyretin. As we know, the changes of transthyretin concentration is regarded as one of the important reasons that metabolic or biochemical abnormity in thyroid gland responsible for hepatopathy[33,34]. The up-regulations of transthyretin in liver tissue is speculated to have following functions: (1) breaking the balance of HSS (hepatocyte stimulate substrate) and inhibitor and thus enhancing the dedifferention. We have cloned two up-regulated genes, one of which is 78.6% homologous to rat transferring gene and the other is 72% homologous to mouse embryonic stem cell gene from 112 hr cDNA library following SISPH (remains to be published), It is also known that over-sexpression of transferring gene and embryonic stem cell gene have close relations with the cellular dedifferentiation; (2) influenceing the induction of enzyme and consumption of oxygen. Compensatory respiration was promoted in mitochondrion responsible for partial hypoxia following PH, but the energy produced from tricarboxyic acid cycle couldn't meet the structural recovery of damaged liver due to successive partial hepatectomy and thus triggered the related genes to express in hypoxia metabolism. The over-expressed gene of alcohol dehydrogenase have cloned from 112 hr cDNA library following SISPH (remains to be published) seems to explain this hypothesis which is consistent with the result of Cheng Lingzhong.
In conclusion, complicated physiological and biochemical reaction changed in the liver tissue following SISPH, we postulated that this kind of up-regulated gene LR1 expression involve in liver regeneration may (1) relate with compensated increase of stress molecules following SISPH; (2) lead to the initiation of other metabolism; (3) maintain the homeostasis in the body. As to, whether severe dedifferentiation happened following SISPH and what are the key dedifferential factors and how they influence the dedifferentiation, remain to be further investigated.