Published online Feb 15, 2000. doi: 10.3748/wjg.v6.i1.107
Revised: September 22, 1999
Accepted: October 12, 1999
Published online: February 15, 2000
AIM: To study the genetic susceptibility of HLA-DQA1 alleles to duodenal ulcer in Wuhan Hans. METHODS: Seventy patients with duodenal ulcer and fifty health y controls were examined for HLA-DQA1 genotypes. HLA-DQA1 typing was carried out by digesting the locus specific polymerase chain reaction amplified products with alleles specific restriction enzymes (PCR-RFLP), i.e. Apal I, Bsaj I, Hph I, Fok I, Mbo II and Mnl I.
RESULTS: The allele frequencies of DQA1*0301 and DQA1*0102 in patients with duodenal ulcer were significantly higher and lower respectivel y than those in healthy controls (0.40 vs 0.20, P = 0.003, Pc orret = 0.024) and (0.05 vs 0.14, P = 0.012, but P corret > 0.05), respectively.
CONCLUSION: DQA1*0301 is a susceptible gene for duodenal ulcer in Wuhan Hans, and there are immunogenetic differences in HLA-DQA1 locus between duodenal ulcer patients and healthy controls.
- Citation: Du YP, Deng CS, Lu DY, Huang MF, Guo SF, Hou W. Relation between HLA-DQA1 genes and genetic susceptibility to duodenal ulcer in Wuhan Hans. World J Gastroenterol 2000; 6(1): 107-110
- URL: https://www.wjgnet.com/1007-9327/full/v6/i1/107.htm
- DOI: https://dx.doi.org/10.3748/wjg.v6.i1.107
Duodenal ulcer is a set of extensive heterogenous hereditary diseases with varied pathogeny and pathogenesis[1], there were some immunological changes in some patients with duodenal ulcer[2-4]. Immunopathogenesis may play a role in the pathogenesis of duodenal ulcer. Human leukocyte antigen (HLA-DQA1 gene) contributes to the pathogenesis of some diseases. Investigating the correlation between HLA-DQA1 gene and duodenal ulcer may afford important clues to reveal duodenal ulcer pathogenesis. HLA-DQA1 genotypes are highly polymorphism and can not be determined by ordinary serological methods. We applied polymerase chain reaction-restricted fragment length polymorphism (PCR-RFLP) nucleotide typing technique to study the genetic susceptibility of HLA-DQA1 alleles to duodenal ulcer in Wuhan Hans.
We examined 70 patients with duodenal ulcer (53 male, 17 female; mean age 38 years) and 50 healthy controls (38 male, 12 female; mean age 36 years) for HLA-DQA1 genotypes. All these subjects were from the Digestive Department, Second Hospital of Hubei Medical University. Duodenal ulcer was diagnos ed endoscopically. The ulcerogenic drugs, such as aspirin, indomethacin or corti costeroids were ruled out as the etiological factors in all of the duodenal ulcer patients. Patients with combined duodenal and gastric ulcers were excluded from this study. All patients and controls were Chinese Hans living in Wuhan.
HLA-DQA1 locus specific PCR primers were designed by Ota M[5]. Primer GH26: 5’-GTGCTGCAGGTGTAAACTTGTACCAG-3’, primer GH27: 5’-CACGGATCCGGTAGCAGCGGTAGAGTTG-3’, they were synthesized by Shanghai Branch, Canadian Sangon Company. Alleles specific restriction enzymes (Apal I, Bsaj I, Hph I, Fok I, Mbo II and Mnl I) were all purchased from New England Biolabs Company. Taq DNA polyenzyme and DNA dNTP were purchased from Canadian Sangon Company. Proteinase K and pBR322/Hae III Mark were provided by Huamei Bioengineer Company.
DNA extraction Leukocytes from anticoagulated whole blood were isolated with hypo-osmotic haemolytic method. DNA was extracted by phenol-chl oroform extraction method from leukocytes.
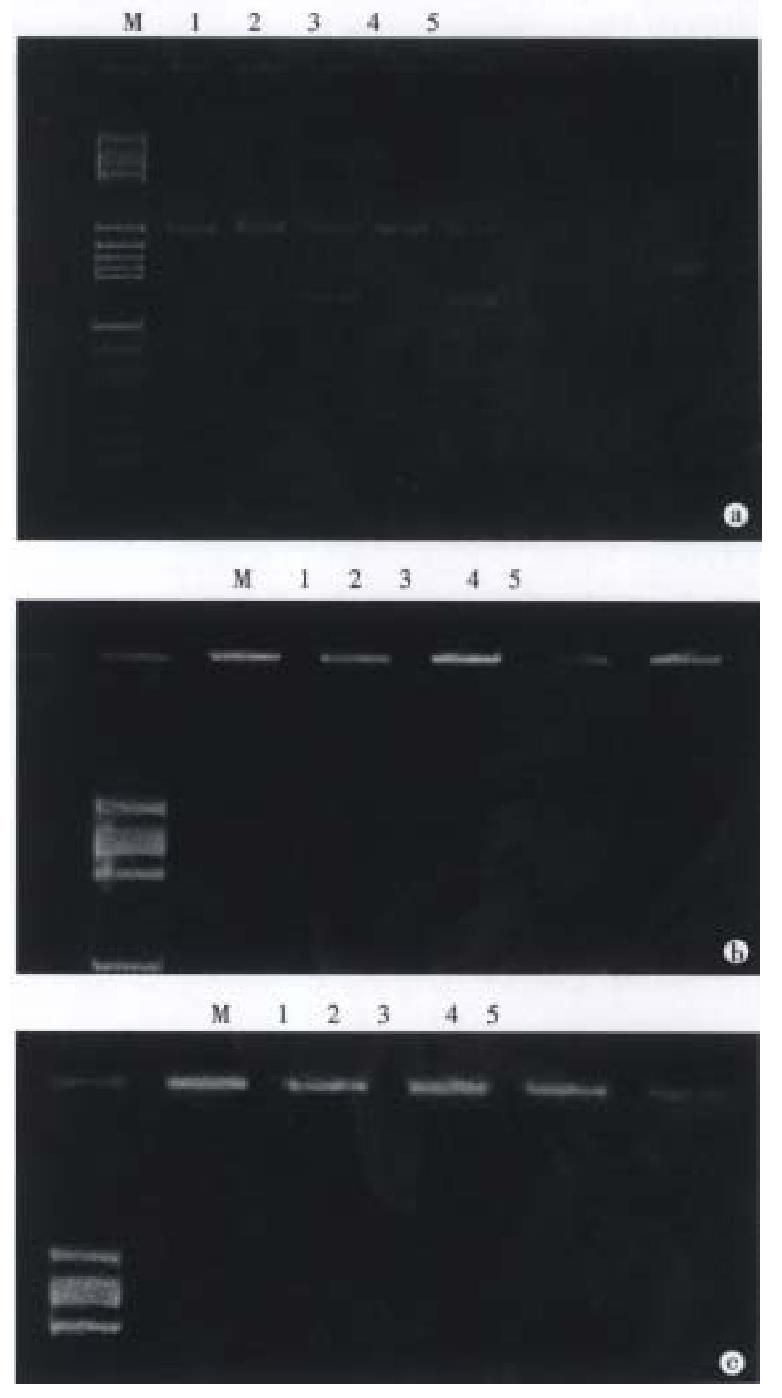
PCR amplification A total amount of 100 μL reaction soluti on contained 1 μg DNA sample, 200 μmol/L dNTP and 50 pmol each of primers 1 and 2. The amplification of HLA-DQA1 exon 2 was carried out in one reaction system and PCR procedure was as follows: predenaturation at 94 °C for 5 min, addition of 2.5 μ Taq DNA polymerase, denaturation at 94 °C for 1 min, annealing at 62 °C for 2 min and extension at 72 °C for 2 min repeated 30 cycles and finally extension at 72 °C for 5 min to terminate the reaction. Because some alleles of HLA-DQA1 locus had a 3 bp deletion in the PCR-amplification the size of the amplified region of the DQA1 exon 2 was 239 bp or 242 bp as indicated (Figure 1).
Digestion with restriction endonucleases After amplification, aliquots (7 μL) of the reaction mixture were digested with restriction endonucleases (Apal-I, Hph-I, Fok-I and Mbo-II: 5 units) at 37 °C for 1 h after addition of appropriate incubation buffer. When digested by Bsaj-I, the reaction mixture was incubated at 60 °C for 1 h.
Acrylamide gel electrophoresis Samples of the restriction enzym e-cleaved product amplified DNA were usually subjected to electrophoresis in 12% polyacrylamide gel in a horizontal minigel apparatus. They were expected to allow discrimination of seven alleles among the eight DQA1 alleles because the DQA1*0101 and DQA1*0102 alleles gave the same restriction map. However, when digested with Mnl-I enzyme, DQA1*0101 and DQA1*0102 can be discriminate d with 15% polyacrylamine gel. Cleavage or no cleavage of amplified fragments was detected by staining with ethidium bromide. Eight alleles of HLA-DQA1 locus were determined by this method[5]. The restriction map of the DQA1 alleles concerning these enzymes is shown in Figure 2.
Chi-square test or Yate’s correction Allele detect frequencies (f) were made by the direct count. According to Hardy-Weinbrg theorem, gene frequencies (AF) were calculated by AF = 1-1-f. Comparisons of allele frequencies were made by the exact probabilities in 2 × 2 table P test. Relative risk frequencies (RR) were made by Wolf formula. The corrected P value was P times the number of alleles in comparison[6].
The distribution of HLA-DQA1 alleles is shown in Table 1. DQA1*0301 is the allele commonly seen in healthy controls. The detect frequency and the allele frequency of DQA1*0301 was 0.36 and 0.20 in healthy controls, and 0.64 and 0.40 in patients with duodenal ulcer, respectively. The increased allele freque ncy of DQA1*0301 in patients with duodenal ulcer (0.40) was statistically si gnificant (RR = 3.20, P = 0.003, Pcorrect = 0.024) compared with healthy controls (0.20). In contrast, the detect frequency and the alle le frequency of DQA1*0102 was 0.26 and 0.14 in healthy controls, and 0.09 and 0.05 in patients with duodenal ulcer, respectively. The decreased frequency of DQA1*0102 in patients with duodenal ulcer (0.05) controls (0.14) was statistically significant compared with healthy (RR = 0.27, P = 0.012 , but Pcorrect > 0.05). There were no significant differences in other allele frequencies between duodenal ulcer patients and healthy controls.
Various factors are accepted as the causes of duodenal ulcer disease. Not only inherited factor, but also immuno-dysfunction is associated with duodenal ulcer. Since the discovery of -Helicobacter pylori (H. pylori)-its infection has been widely accepted as the predominant cause of duodenal ulcer disease. T he pathogenic mechanisms that H. pylori causes human disease remain poorly understood. The human leukocyte antigen (HLA) DQA1 gene contributes to the host response against H. pylori. Many immune responses are controlled by genes of the major histocompatibility complex which encode HLA. Individuals with different HLA types may differ in susceptibility or resistance to particular infectious pathogens, and associations between HLA polymorphism and susceptibility or resistance to in fectious or autoimmune diseases have been identified[7,8]. The polymorphic HLA-DQA1 genes encode polypeptides which fold together to form a receptor that specifies T-lymphocyte recognition of self and foreign peptides[9]. Therefore we examined the HLA DQA1 locus in patients with duodenal ulcer in an attempt to investigate immunogenetic differences in the host.
Researches in the correlation between HLA and peptic ulcer disease were started sixty years ago. The finding showed that HLA-B5 antigen was associated with pepticulcer[10-13]. However, traditional serological method was used in some investigations, but it was obsolete and inaccurate. The individual genetic difference of HLA at the level of DNA was produced by encoded gene [14]. To understand the disease essence, correlation between peptic ulcer and HLA should be further studied with nucleotide typing technique.
DQA1 gene is the most polymorphism in HLA Class II gene and is strongly associated with some diseases. We analysed nucleotide sequences of PCR-amplified regions in the DQA1 genes for allele specific restriction sites to study DQA1 genes involved in the genetic susceptibility to duodenal ulcer in Wuhan Hans. The result of the present study indicated a significant difference in the frequencies of HLA-DQA1 alleles between duodenal ulcer patients and healthy controls. The allele frequency of DQA1*0301 was significantly higher (RR = 3.20, P = 0.003, Pcorret = 0.024) in duodenal ulcer patients (0.40) than in healthy controls (0.20). In contrast, the allele frequency of DQA1*0102 was significantly lower (RR = 0.27, P = 0.012) in duodenal ulcer patients (0.05) than in healthy controls (0.14), but Pcorret > 0.05. These results suggest that the HLA-DQA1*0301 may contribute to the susceptibility to
duodenal ulcer.
DQA1 chain is not on isolated entity and is tightly linked with DR and DQB locus genes in the all genes region of the major histocompatibility complex. The result of the present study may provide a clue that HLAII genes are more strongly associated with duodenal ulcer than HLAI genes. The clue could not affirm which locus gene of HLAII Class genes is more important. Further genetic analysis of DRB1 and DQB1 genes, using various DNA markers to clarify the host genetic factors of susceptibility or resistance to duodenal ulcer, is required.
Edited by You DY and Ma JY
| 1. | Lam SK, Hui WM, Shiu LP, Ng MM. Society stress and peptic ulcer perforation. J Gastroenterol Hepatol. 1995;10:570-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 2. | De Lazzari F, Mancin O, Plebani M, Venturi C, Battaglia G, Vianello F, Galliani EA, Di Mario F, Naccarato R. High IgE serum levels and "peptic" ulcers: clinical and functional approach. Ital J Gastroenterol. 1994;26:7-11. [PubMed] [Cited in This Article: ] |
| 3. | Negrini R, Lisato L, Zanella I, Cavazzini L, Gullini S, Villanacci V, Poiesi C, Albertini A, Ghielmi S. Helicobacter pylori infection induces antibodies cross-reacting with human gastric mucosa. Gastroenterology. 1991;101:437-445. [PubMed] [Cited in This Article: ] |
| 4. | Gionchetti P, Vaira D, Campieri M, Holton J, Menegatti M, Belluzzi A, Bertinelli E, Ferretti M, Brignola C, Miglioli M. Enhanced mucosal interleukin-6 and -8 in Helicobacter pylori-positive dyspeptic patients. Am J Gastroenterol. 1994;89:883-887. [PubMed] [Cited in This Article: ] |
| 5. | Ota M, Seki T, Nomura N, Sugimura K, Mizuki N, Fukushima H, Tsuji K, Inoko H. Modified PCR-RFLP method for HLA-DPB1 and -DQA1 genotyping. Tissue Antigens. 1991;38:60-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 136] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 6. | Zhao TM. Correlation between HLA and Disease. See: Zhao TM Editor in Chief Classification and Application of HLA 1st edition. Shanghai Scientific Technology Publishing Company. 1984;187-220. [Cited in This Article: ] |
| 7. | Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595-600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1071] [Cited by in F6Publishing: 980] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 8. | Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599-604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1542] [Cited by in F6Publishing: 1493] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 9. | Pullen AM, Marrack P, Kappler JW. The T-cell repertoire is heavily influenced by tolerance to polymorphic self-antigens. Nature. 1988;335:796-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 325] [Cited by in F6Publishing: 410] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 10. | Rotter JI, Rimoin DL, Gursky JM, Terasaki P, Sturdevant RA. HLA-B5 associated with duodenal ulcer. Gastroenterology. 1977;73:438-440. [PubMed] [Cited in This Article: ] |
| 11. | O'Brien BD, Thomson AB, Dossetor JB. HLA and peptic ulcer. Dig Dis Sci. 1979;24:314-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 11] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 12. | Potashov LV, Savranskiĭ VM, Berkos AS, Bubnova LN, Morozov VP. [The association of HLA antigens with gastric and duodenal peptic ulcer]. Vestn Khir Im I I Grek. 1994;152:14-17. [PubMed] [Cited in This Article: ] |
| 13. | Wang LX, Chen PS, Zheng ZT. [Study on the association of HLA with peptic ulcer]. Zhonghua Neike Zazhi. 1986;25:409-11, 445. [PubMed] [Cited in This Article: ] |
| 14. | Benacerraf B. Role of MHC gene products in immune regulation. Science. 1981;212:1229-1238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 483] [Cited by in F6Publishing: 489] [Article Influence: 11.4] [Reference Citation Analysis (0)] |