Copyright
©The Author(s) 2025.
World J Gastroenterol. Feb 21, 2025; 31(7): 101104
Published online Feb 21, 2025. doi: 10.3748/wjg.v31.i7.101104
Published online Feb 21, 2025. doi: 10.3748/wjg.v31.i7.101104
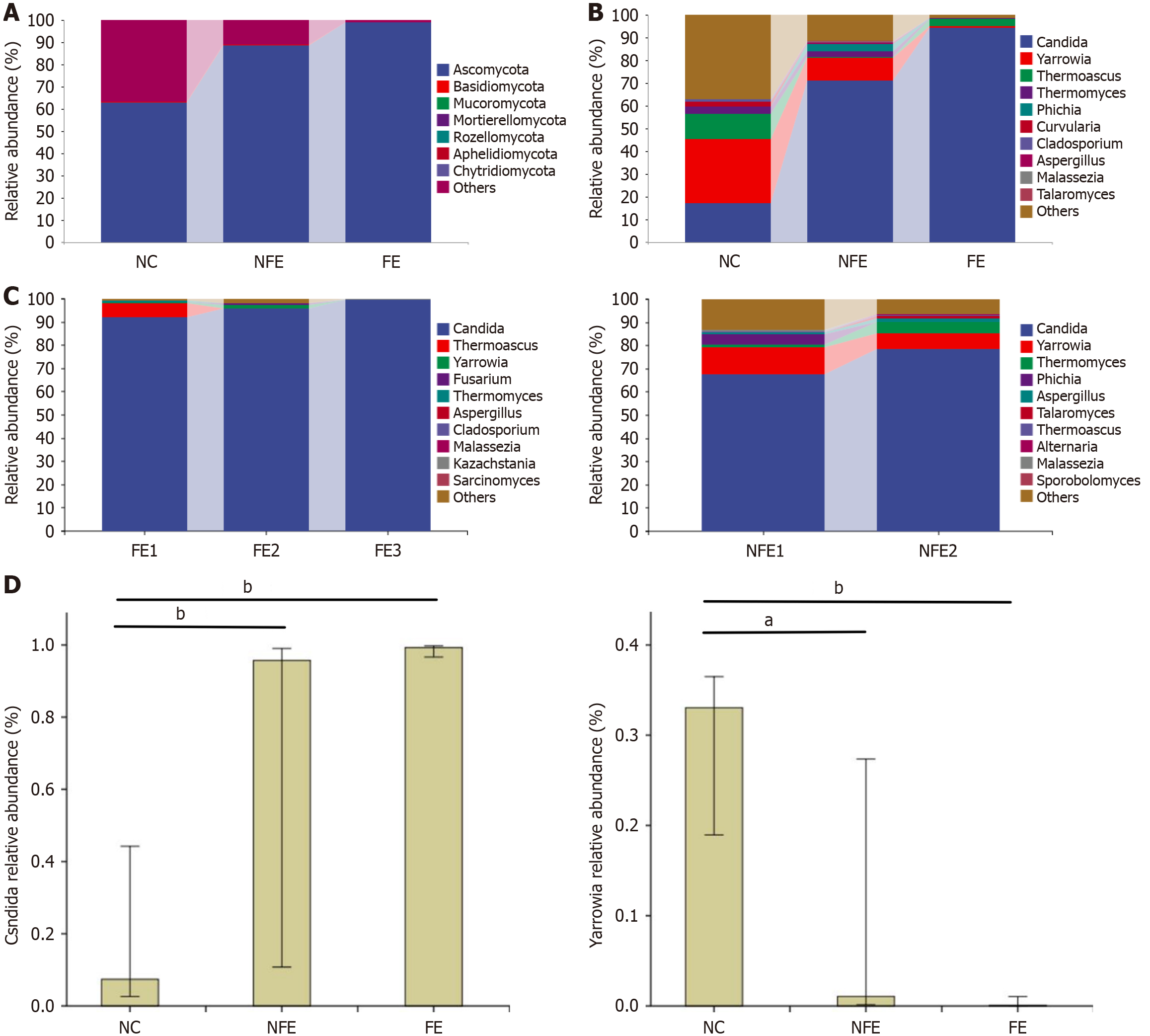
Figure 3 Taxonomic characteristics of esophageal fungi microflora in the esophageal normal control group, suspected fungal esophagitis group, and fungal esophagitis group.
A: Relative abundance at the phylum level in three groups; B: Relative abundance at the genus level in three groups; C: Relative abundance at the genus level of the Kodsi subgroup in the suspected fungal esophagitis (NFE) and FE groups; D: Relative abundance of Candida genera and Yarrowia genera in three groups. Diverse letters specify statistically significant differences among groups. aP < 0.05, bP < 0.001. NC: The esophageal normal control; NFE: Suspected fungal esophagitis; FE: Fungal esophagitis.
- Citation: Song YK, Zheng L, Liu AX, Ma JJ. Internal transcribed spacer sequencing to explore the intrinsic composition of fungal communities in fungal esophagitis. World J Gastroenterol 2025; 31(7): 101104
- URL: https://www.wjgnet.com/1007-9327/full/v31/i7/101104.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i7.101104