Copyright
©The Author(s) 2025.
World J Gastroenterol. Jan 21, 2025; 31(3): 95207
Published online Jan 21, 2025. doi: 10.3748/wjg.v31.i3.95207
Published online Jan 21, 2025. doi: 10.3748/wjg.v31.i3.95207
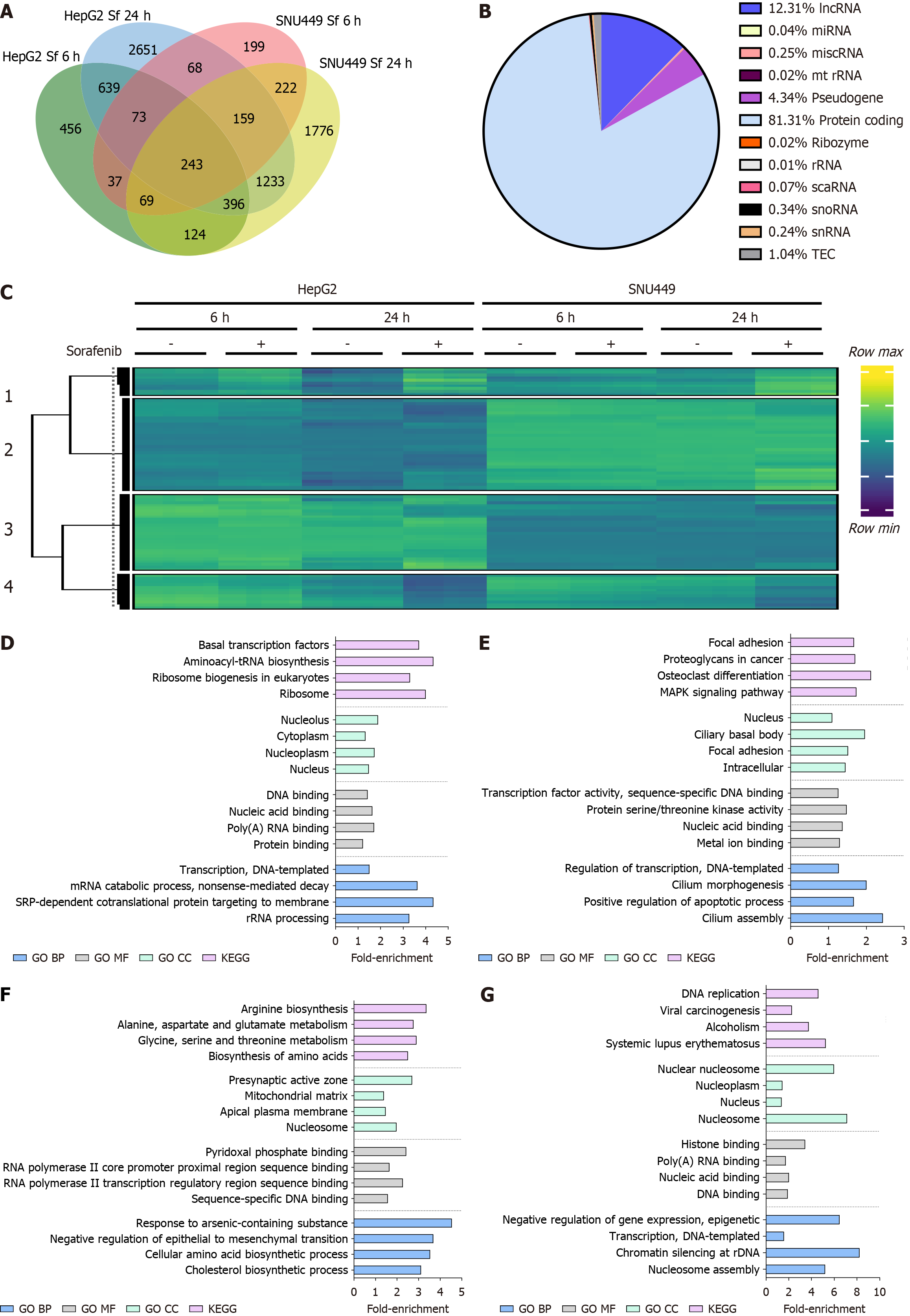
Figure 1 Expression profile of long RNA species and enrichment analysis.
A: Venn diagram of differentially expressed (DE) genes in HepG2 and SNU449 cells treated with Sorafenib for 6 and 24 hours; B: Pie chart showing identification of RNA species; C: Heatmap of the 8345 DE genes grouped into 4 row clusters; D: Top 4 Gene Ontology terms and Kyoto Encyclopedia of Genes and Genome pathways in cluster 1; E: Cluster 2; F: Cluster 3; G: Cluster 4 identified in the heatmap.
- Citation: de la Cruz-Ojeda P, Parras-Martínez E, Rey-Pérez R, Muntané J. In silico analysis of lncRNA-miRNA-mRNA signatures related to Sorafenib effectiveness in liver cancer cells. World J Gastroenterol 2025; 31(3): 95207
- URL: https://www.wjgnet.com/1007-9327/full/v31/i3/95207.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i3.95207