Copyright
©The Author(s) 2025.
World J Gastroenterol. Apr 28, 2025; 31(16): 105985
Published online Apr 28, 2025. doi: 10.3748/wjg.v31.i16.105985
Published online Apr 28, 2025. doi: 10.3748/wjg.v31.i16.105985
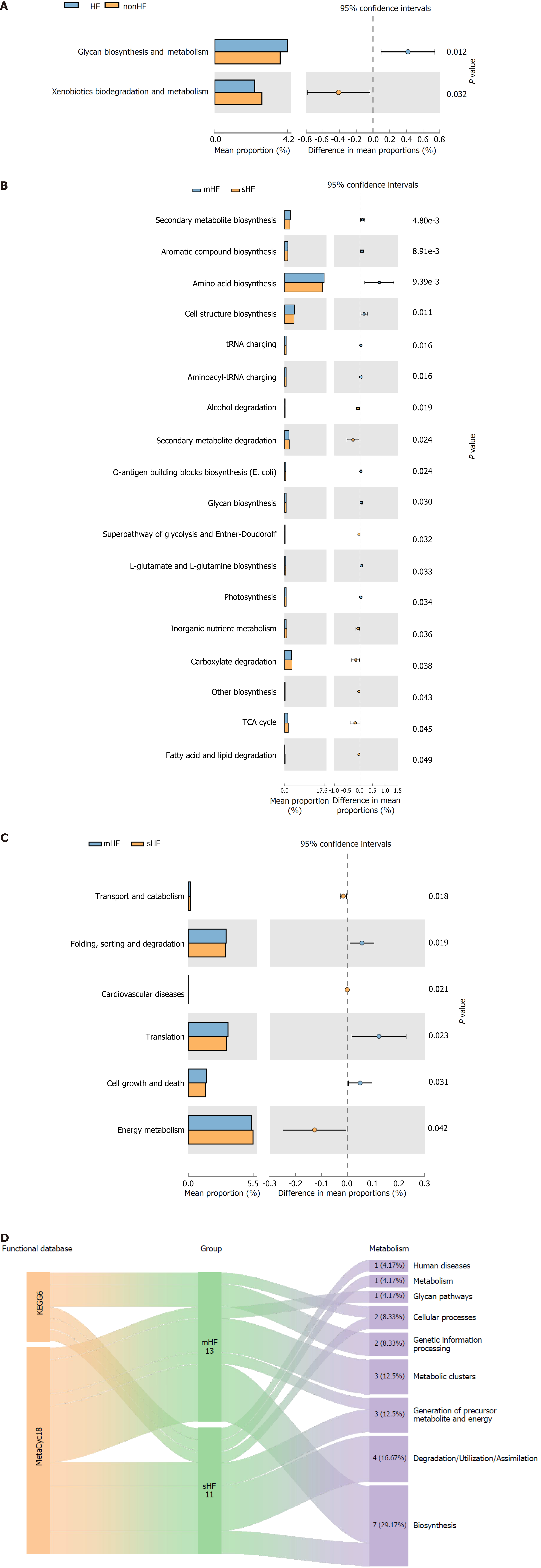
Figure 6 The relationship between key signaling pathways and hepatic fibrosis progression.
A: The differential microbiota metabolic pathways between non-hepatic fibrosis (HF) and HF groups in Kyoto Encyclopedia of Genes and Genomes (KEGG) databases; B and C: The differential microbiota metabolic pathways between mild HF (mHF) and significant HF (sHF) groups in KEGG and MetaCyc databases, respectively; D: The subgroup analysis of mHF and sHF revealed 24 differential pathways in KEGG and MetaCyc databases. t-test was used to assess the significance of differences in microbial community functioning between different subgroups. HF: Hepatic fibrosis; mHF: Mild hepatic fibrosis; sHF: Significant hepatic fibrosis; TCA: Tricarboxylic acid; KEGG: Kyoto Encyclopedia of Genes and Genomes.
- Citation: Zhu Y, Geng SY, Chen Y, Ru QJ, Zheng Y, Jiang N, Zhu FY, Zhang YS. Machine learning algorithms reveal gut microbiota signatures associated with chronic hepatitis B-related hepatic fibrosis. World J Gastroenterol 2025; 31(16): 105985
- URL: https://www.wjgnet.com/1007-9327/full/v31/i16/105985.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i16.105985