Copyright
©The Author(s) 2024.
World J Gastroenterol. Feb 28, 2024; 30(8): 919-942
Published online Feb 28, 2024. doi: 10.3748/wjg.v30.i8.919
Published online Feb 28, 2024. doi: 10.3748/wjg.v30.i8.919
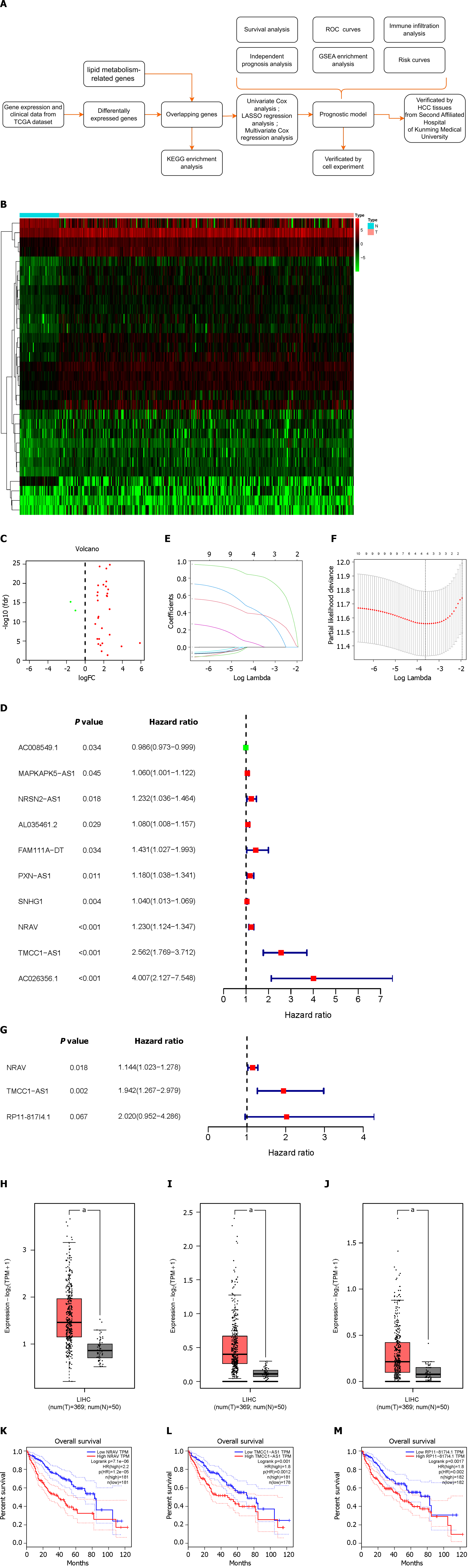
Figure 1 Differentially expressed lipid metabolism-related and prognosis-related long non-coding RNAs.
A: Flow chart of the analysis process in our study; B and C: Heatmap and volcano plot demonstrating differentially expressed lipid metabolism-related long non-coding RNAs (lncRNAs) between Hepatocellular carcinoma (HCC) tissues and nontumor tissues. Red, positive regulation; green, negative regulation; D: Forest plot of hazard ratios (HR) constrating the prognostic value of lncRNAs. The dashed line indicates that the location of the HR was 1; E: Construction and analysis of the prognostic risk model. Plots for Least absolute shrinkage and selection operator expression coefficients of 10 Lipid metabolism-related lncRNAs; F: Cross-validation plot for the penalty term; G: The HRs and P values from the multivariate Cox regression analysis are shown in the forest plot; H-J: Three differentially expressed lncRNAs between HCC and nontumor tissues in the GeneExpression Profiling Interactive Analysis database; K-M: Survival curves of patients with differential expression of three lncRNAs in GEPIA. HCC: Hepatocellular carcinoma; Lasso: Least absolute shrinkage and selection operator; KEGG: Kyoto Encyclopedia of Genes and Genomes; NRAV: Negative regulator of antiviral response; TMCC1-AS1: RNA transmembrane and coiled-coil domain family 1 antisense RNA 1; FC: Fold change; FDR: False discovery rate; TPM: Transcripts Per Kilobase of exon model per Million mapped reads; TCGA: The Cancer Genome Atlas; ROC: Receiver operating characteristic.
- Citation: Wang RY, Yang JL, Xu N, Xu J, Yang SH, Liang DM, Li JZ, Zhu H. Lipid metabolism-related long noncoding RNA RP11-817I4.1 promotes fatty acid synthesis and tumor progression in hepatocellular carcinoma. World J Gastroenterol 2024; 30(8): 919-942
- URL: https://www.wjgnet.com/1007-9327/full/v30/i8/919.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i8.919