Copyright
©The Author(s) 2024.
World J Gastroenterol. Dec 7, 2024; 30(45): 4817-4835
Published online Dec 7, 2024. doi: 10.3748/wjg.v30.i45.4817
Published online Dec 7, 2024. doi: 10.3748/wjg.v30.i45.4817
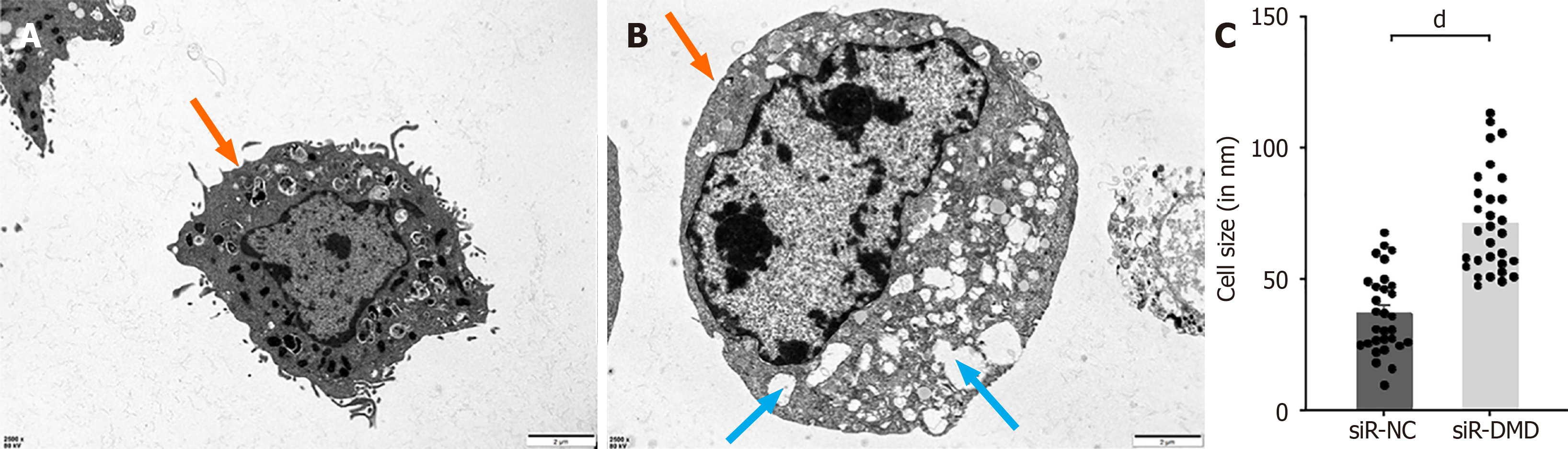
Figure 6 Representative images of two groups of cells examined under a transmission electron microscope.
A: Outline of control group intestinal smooth muscle cells. Orange arrows indicate intact cell membranes. Scale bar = 2 μm; B: Outline of experimental group intestinal smooth muscle cells. Orange arrows indicate intact cell membranes. Blue arrows indicate an abundance of vacuolar-like structures. Scale bar = 2 μm; C: Characterization of cell size under a transmission electron microscope. dP < 0.0001. siR-NC: Negative control small interfering RNA; siR-DMD: Dystrophin small interfering RNA.
- Citation: Li WZ, Xiong Y, Wang TK, Chen YY, Wan SL, Li LY, Xu M, Tong JJ, Qian Q, Jiang CQ, Liu WC. Quantitative proteomics analysis reveals the pathogenesis of obstructed defecation syndrome caused by abnormal expression of dystrophin. World J Gastroenterol 2024; 30(45): 4817-4835
- URL: https://www.wjgnet.com/1007-9327/full/v30/i45/4817.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i45.4817