Copyright
©The Author(s) 2024.
World J Gastroenterol. Nov 7, 2024; 30(41): 4461-4480
Published online Nov 7, 2024. doi: 10.3748/wjg.v30.i41.4461
Published online Nov 7, 2024. doi: 10.3748/wjg.v30.i41.4461
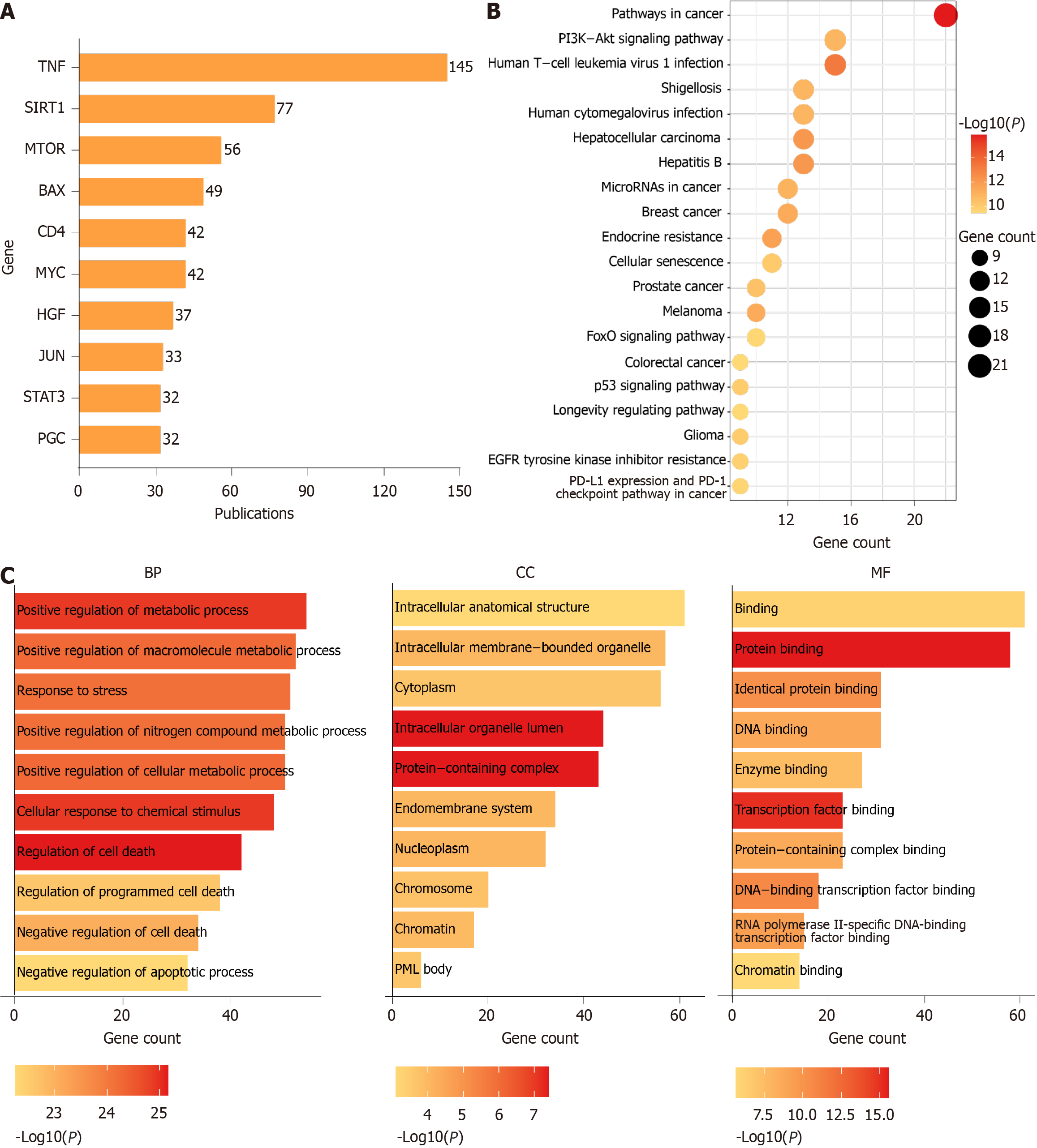
Figure 10 Gene analysis in liver aging research.
A: Top 20 genes reported in publication abstracts; B: Kyoto Encyclopedia of Genes Genomes pathway enrichment analysis of the top 64 genes; C: Gene Ontology enrichment analysis of the top 64 genes. BP: Biological process; MF: Molecular function; CC: Cellular component; TNF: Tumor necrosis factor; SIRT1: Sirtuin 1; mTOR: Mechanistic target of rapamycin; HGF: Hepatocyte growth factor; STAT3: Signal transducer and activator of transcription 3; PGC: Peroxisome proliferator-activated receptor gamma coactivator 1; PI3K: Phosphatidylinositol 3-kinase; Akt: Protein kinase B; FoxO: Forkhead box O; PD-L1: Programmed death-ligand 1; PD-1: Programmed cell death protein-1.
- Citation: Han QH, Huang SM, Wu SS, Luo SS, Lou ZY, Li H, Yang YM, Zhang Q, Shao JM, Zhu LJ. Mapping the evolution of liver aging research: A bibliometric analysis. World J Gastroenterol 2024; 30(41): 4461-4480
- URL: https://www.wjgnet.com/1007-9327/full/v30/i41/4461.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i41.4461