Copyright
©The Author(s) 2024.
World J Gastroenterol. Sep 14, 2024; 30(34): 3894-3925
Published online Sep 14, 2024. doi: 10.3748/wjg.v30.i34.3894
Published online Sep 14, 2024. doi: 10.3748/wjg.v30.i34.3894
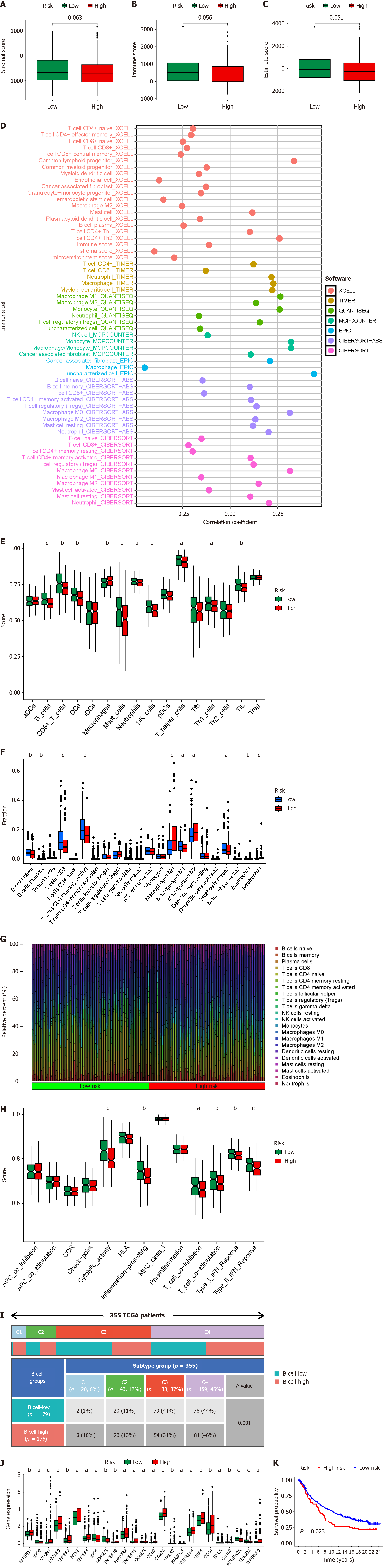
Figure 8 Assessment of the immune microenvironment in different risk score groups.
A: Box plot illustrates the stromal score across different risk score groups; B: Box plot displays the immune score across different risk score groups; C: Box plot showcases the estimate score across different risk score groups; D: Bubble plot demonstrates the correlation between risk score and immune cell infiltration within hepatocellular carcinoma samples; E: Box plot shows the abundance of immune cells across different risk score groups calculated based on the single-sample gene set enrichment analysis (ssGSEA) algorithm; F: Box plot exhibits the abundance of immune cells across different risk score groups calculated based on the cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT) algorithm; G: Heatmap displays the distribution and proportions of immune cells across different risk score groups calculated based on the CIBERSORT algorithm; H: Box plot demonstrates the immune functional status across different risk score groups calculated based on the ssGSEA algorithm; I: Association between The Cancer Genome Atlas immune subtypes and BRGs prognostic subtypes; J: Box plot showcases the expression of immune checkpoint genes across different risk score groups; K: Survival analysis depicts the immune therapy response and efficacy in different risk score groups of the IMvigor210 immune therapy cohort. aP < 0.05; bP < 0.01; cP < 0.001. LIHC: Liver hepatocellular carcinoma; CD: Cluster of differentiation; NK: Nature kill; DC: Dendritic cells; HLA: Human leukocyte antigen; TCGA: The cancer genome atlas.
- Citation: Xu KQ, Gong Z, Yang JL, Xia CQ, Zhao JY, Chen X. B-cell-specific signatures reveal novel immunophenotyping and therapeutic targets for hepatocellular carcinoma. World J Gastroenterol 2024; 30(34): 3894-3925
- URL: https://www.wjgnet.com/1007-9327/full/v30/i34/3894.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i34.3894