Copyright
©The Author(s) 2024.
World J Gastroenterol. Jun 28, 2024; 30(24): 3086-3105
Published online Jun 28, 2024. doi: 10.3748/wjg.v30.i24.3086
Published online Jun 28, 2024. doi: 10.3748/wjg.v30.i24.3086
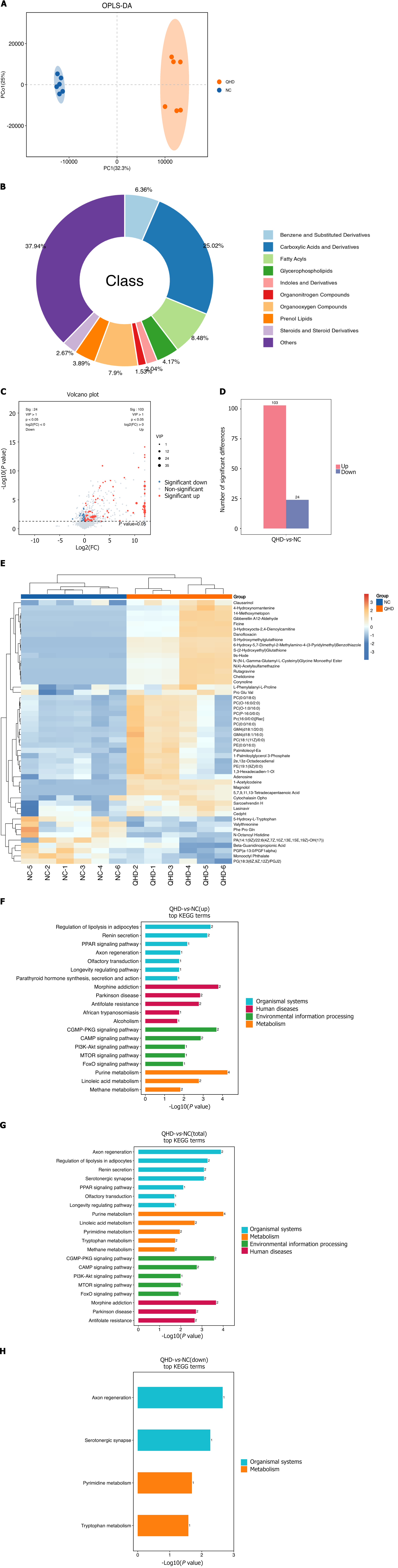
Figure 7 Metabolic difference analysis and metabolomic pathway enrichment analysis for Qingre Huashi decoction vs NC.
A: Classification of metabolites; B: OPLS-DA analysis revealed that the samples from the NC group and Qingre Huashi decoction (QHD) group exhibited distinct clustering patterns; C: Number of differential metabolites between the QHD group and NC group; D: QHD vs NC. Red dots represent significantly upregulated differential metabolites [P < 0.05, variable importance in projection (VIP) > 1 and FC > 1], blue dots refer to significantly downregulated differential metabolites (P < 0.05, VIP > 1 and FC < 1), and gray dots stand for metabolites with no significant difference. Each dot in the plot represents a metabolite, with the x-axis showing the log2 (FC) values of the comparison between the two groups, and the y-axis representing the -log10 (P value) values; E: Hierarchical clustering of all significantly differentially expressed metabolites and top 50 differentially expressed metabolites ranked by VIP. The horizontal axis represents sample names, while the vertical axis indicates differential metabolites. The color gradient from blue to red indicates the abundance of metabolite expression, with red showing higher expression levels of differential metabolites; F-H: KEGG pathway enrichment analysis on differentially expressed metabolites. The x-axis represents the -log10 P-value for each pathway, the y-axis stands for different pathway names, and the numbers on the bars indicate the number of differentially expressed metabolites annotated to that pathway.
- Citation: Lin MM, Yang SS, Huang QY, Cui GH, Jia XF, Yang Y, Shi ZM, Ye H, Zhang XZ. Effect and mechanism of Qingre Huashi decoction on drug-resistant Helicobacter pylori. World J Gastroenterol 2024; 30(24): 3086-3105
- URL: https://www.wjgnet.com/1007-9327/full/v30/i24/3086.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i24.3086