Copyright
©The Author(s) 2023.
World J Gastroenterol. Nov 28, 2023; 29(44): 5872-5881
Published online Nov 28, 2023. doi: 10.3748/wjg.v29.i44.5872
Published online Nov 28, 2023. doi: 10.3748/wjg.v29.i44.5872
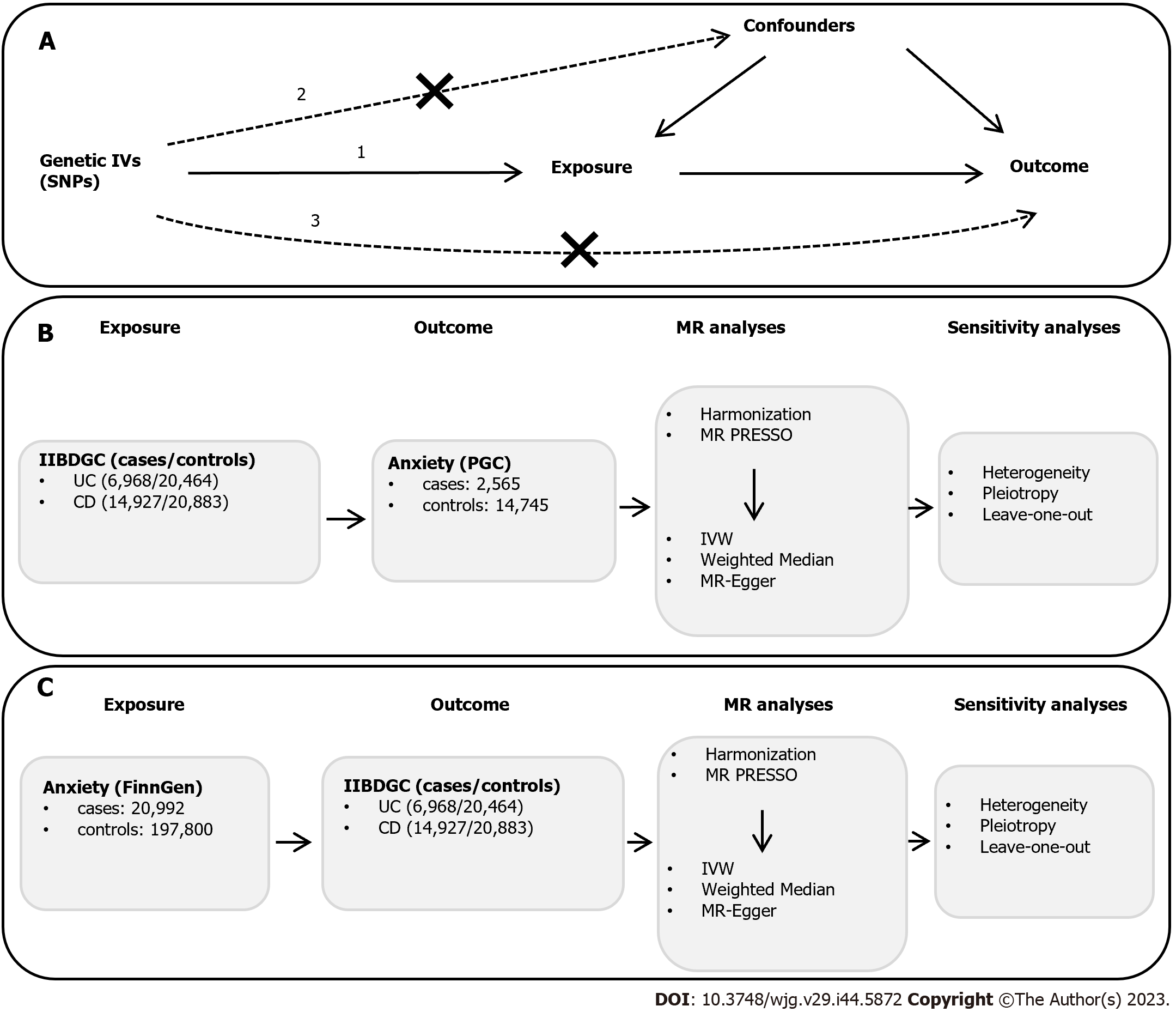
Figure 1 Schematic overview of the study design.
A: Mendelian randomization (MR) analysis illustration. There are three principal assumptions in MR design: (1) Relevance assumption: the genetic instrument variants (IVs) are strongly associated with the exposure; (2) independence assumption: the genetic IVs do not affect the outcome through the confounders; and (3) exclusion assumption: The genetic IVs do not affect the outcome directly but only via indirect exposure; B: MR analysis from ulcerative colitis (UC) or Crohn’s disease (CD) to anxiety. UC- or CD-related single-nucleotide polymorphisms (SNPs) obtained from genome-wide association studies (GWAS) statistics in the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) consortium were identified as IVs, and GWAS summary statistics of anxiety were retrieved from Psychiatric Genomics Consortium; C: MR analysis from anxiety to UC or CD. Anxiety-related SNPs retrieved from FinnGen were identified as IVs, and GWAS statistics for UC and CD were obtained from the IIBDGC consortium. MR analysis was performed after the harmonization process and MR Pleiotropy Residual Sum and Outlier test and subsequently sensitivity analyses to strengthen the MR estimates. MR: Mendelian randomization; IVs: Instrumental variables; UC: Ulcerative colitis; CD: Crohn’s disease; PGC: Psychiatric Genomics Consortium; IVW: Inverse-variance weighted; PRESSO: Pleiotropy Residual Sum and Outlier; IIBDGC: International Inflammatory Bowel Disease Genetics Consortium.
- Citation: He Y, Chen CL, He J, Liu SD. Causal associations between inflammatory bowel disease and anxiety: A bidirectional Mendelian randomization study. World J Gastroenterol 2023; 29(44): 5872-5881
- URL: https://www.wjgnet.com/1007-9327/full/v29/i44/5872.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i44.5872