Copyright
©The Author(s) 2023.
World J Gastroenterol. Jul 7, 2023; 29(25): 4053-4071
Published online Jul 7, 2023. doi: 10.3748/wjg.v29.i25.4053
Published online Jul 7, 2023. doi: 10.3748/wjg.v29.i25.4053
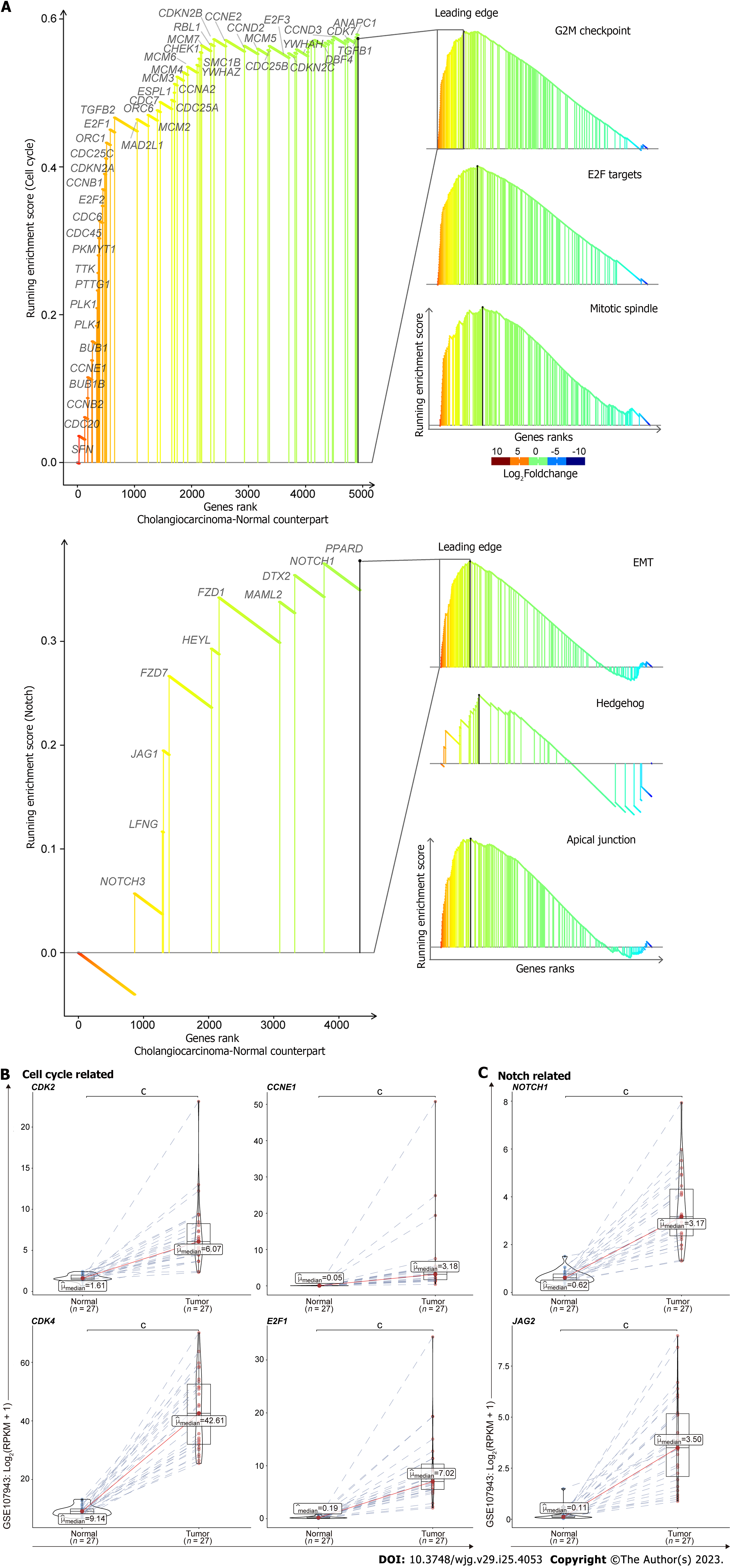
Figure 6 The generalizable oncogenic traits of cell cycle and Notch associated pathways in cholangiocarcinoma.
GSEA identified cell cycle and Notch associated pathways significantly activated in tumors compared to the corresponding adjacent normal livers from GEO database (GSE107943) (A). Upper: Cell cycle associated pathways, including G2M checkpoint, E2F Targets, and Mitotic spindle gene sets; lower: Notch associated pathways, including epithelial mesenchymal transition, Hedgehog, and apical junction gene sets. The mRNA expression levels from the leading edges of cell cycle (B) and Notch (C) associated pathways were analyzed in CCA samples compared to the corresponding adjacent normal livers (pairs = 27). RPKM: Reads per kilobase million; Red: Intrahepatic cholangiocarcinoma; Blue: Para-cancerous normal liver; cP < 0.001 when compared with the relevant normal control.
- Citation: Liu D, Shi Y, Chen H, Nisar MA, Jabara N, Langwinski N, Mattson S, Nagaoka K, Bai X, Lu S, Huang CK. Molecular profiling reveals potential targets in cholangiocarcinoma. World J Gastroenterol 2023; 29(25): 4053-4071
- URL: https://www.wjgnet.com/1007-9327/full/v29/i25/4053.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i25.4053