Copyright
©The Author(s) 2023.
World J Gastroenterol. Mar 21, 2023; 29(11): 1721-1734
Published online Mar 21, 2023. doi: 10.3748/wjg.v29.i11.1721
Published online Mar 21, 2023. doi: 10.3748/wjg.v29.i11.1721
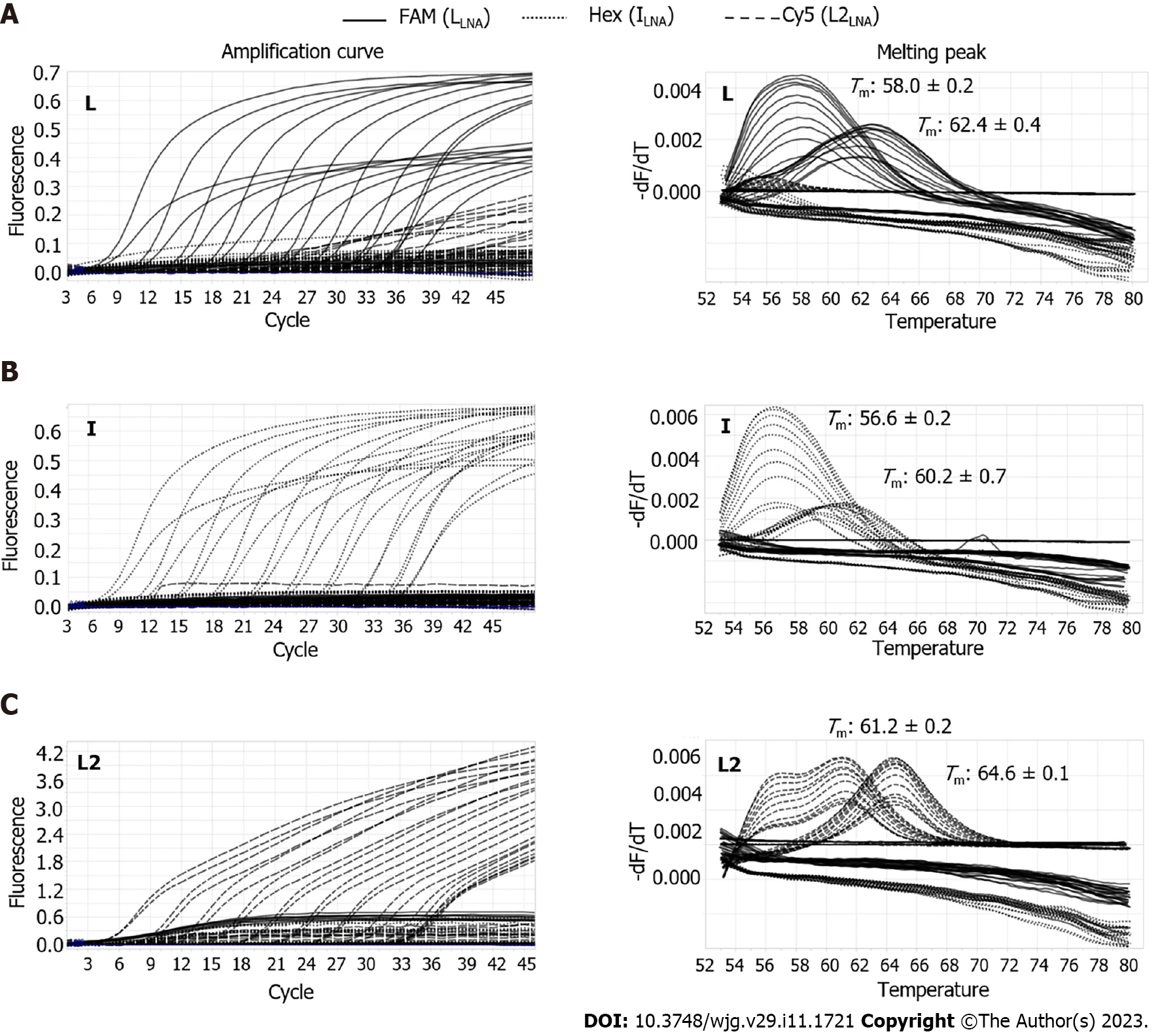
Figure 3 Multiprobe locked nucleic acid real-time polymerase chain reaction for discrimination among three types of polymorphisms in the rt269 codon.
Amplification curves are shown on the left, and melting peaks are shown on the right. A: With L1 wild-type DNA templates, L1-type specific signals in the FAM channel (solid) were detected, showing their dominant amplification and distinct melting temperatures (Tm), with minimal cross signals of amplification and melting peaks generated by weak cross hybridizations of the other probes (I and L2), which were differentiated from the Tm values for I and L2 detection; B: For I variant-type DNA templates, I-type specific signals in the Hex channel (dotted) were detected, showing their exclusive amplifications and distinct Tm values, with no cross signals; C: For L2 variant-type DNA templates, amplification curves showed weak cross signals, but melting peaks were distinct with no cross signals.
- Citation: Kim K, Choi YM, Kim DH, Jang J, Choe WH, Kim BJ. Locked nucleic acid real-time polymerase chain reaction method identifying two polymorphisms of hepatitis B virus genotype C2 infections, rt269L and rt269I. World J Gastroenterol 2023; 29(11): 1721-1734
- URL: https://www.wjgnet.com/1007-9327/full/v29/i11/1721.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i11.1721