Published online Oct 7, 2022. doi: 10.3748/wjg.v28.i37.5483
Peer-review started: June 2, 2022
First decision: August 1, 2022
Revised: August 9, 2022
Accepted: September 20, 2022
Article in press: September 20, 2022
Published online: October 7, 2022
Upper gastrointestinal endoscopy is critical for esophageal squamous cell carcinoma (ESCC) detection; however, endoscopists require long-term training to avoid missing superficial lesions.
To develop a deep learning computer-assisted diagnosis (CAD) system for endoscopic detection of superficial ESCC and investigate its application value.
We configured the CAD system for white-light and narrow-band imaging modes based on the YOLO v5 algorithm. A total of 4447 images from 837 patients and 1695 images from 323 patients were included in the training and testing datasets, respectively. Two experts and two non-expert endoscopists reviewed the testing dataset independently and with computer assistance. The diagnostic performance was evaluated in terms of the area under the receiver operating characteristic curve, accuracy, sensitivity, and specificity.
The area under the receiver operating characteristics curve, accuracy, sensitivity, and specificity of the CAD system were 0.982 [95% confidence interval (CI): 0.969-0.994], 92.9% (95%CI: 89.5%-95.2%), 91.9% (95%CI: 87.4%-94.9%), and 94.7% (95%CI: 89.0%-97.6%), respectively. The accuracy of CAD was significantly higher than that of non-expert endoscopists (78.3%, P < 0.001 compared with CAD) and comparable to that of expert endoscopists (91.0%, P = 0.129 compared with CAD). After referring to the CAD results, the accuracy of the non-expert endoscopists significantly improved (88.2% vs 78.3%, P < 0.001). Lesions with Paris classification type 0-IIb were more likely to be inaccurately identified by the CAD system.
The diagnostic performance of the CAD system is promising and may assist in improving detectability, particularly for inexperienced endoscopists.
Core Tip: Esophageal squamous cell carcinoma (ESCC) poses a heavy burden to high-risk areas, and screening using upper gastrointestinal endoscopy is an established strategy for early detection and prognosis improvement. However, endoscopic detection of superficial-ESCC can be challenging and depends greatly on operator experience. We developed and validated a novel computer-assisted diagnostic system with a deep neural network algorithm to detect superficial ESCC using upper endoscopy with white-light and narrow-band imaging. The system demonstrated high diagnostic accuracy, which is comparable to that of expert endoscopists. The diagnostic performance of non-expert endoscopists was significantly improved under the assistance of this system.
- Citation: Meng QQ, Gao Y, Lin H, Wang TJ, Zhang YR, Feng J, Li ZS, Xin L, Wang LW. Application of an artificial intelligence system for endoscopic diagnosis of superficial esophageal squamous cell carcinoma. World J Gastroenterol 2022; 28(37): 5483-5493
- URL: https://www.wjgnet.com/1007-9327/full/v28/i37/5483.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i37.5483
Esophageal cancer is the seventh most common and sixth most fatal malignancy[1], and esophageal squamous cell carcinoma (ESCC) is the most prevalent histological type[2]. Screening high-risk po
In recent years, artificial intelligence (AI)-aided endoscopy has garnered attention, and several studies have heralded this technique as a promising tool for improving the detection of early ESCC[9-12]. A systematic review showed that the pooled sensitivity and specificity of AI for ESCC diagnosis were 0.95 and 0.92, respectively, and AI performed better than endoscopists, although the differences were not statistically significant[13]. However, few computer-assisted diagnosis (CAD) systems for ESCC that support WLI and narrow-band imaging (NBI) have been applied in clinical practice. We developed and validated a novel CAD system with a deep neural network algorithm to detect superficial ESCC using upper endoscopy with WLI and NBI. Moreover, we assessed its value for endoscopists with differing experience levels of superficial ESCC detection and investigated the characteristics of lesions that were inaccurately identified by the CAD system.
Static and non-magnified WLI or NBI images used for CAD system training and testing for superficial ESCC were retrospectively retrieved from endoscopic databases of four general hospitals in mainland China between January 2016 and April 2019. For the training dataset, 1503 WLI images and 2100 NBI images from 622 cases of superficial ESCC and high-grade intraepithelial neoplasia (HGIN), and 523 WLI images and 321 NBI images from 215 non-cancerous cases (including normal, esophagitis, submucosal lesion, and leukoplakia) were obtained from the First Affiliated Hospital of Zhengzhou University, Xingtai First Hospital, and Tangyin People's Hospital. For the independent testing dataset, 577 WLI images and 793 NBI images from 209 cases of superficial ESCC and HGIN, and 209 WLI images and 116 NBI images from 114 non-cancerous cases were obtained from Changhai Hospital. All images were captured using an Olympus gastroscope (GIF-H260, GIF-H260Z, GIF-H290, GIF-Q260J, or GIF-HQ290; Olympus, Tokyo, Japan). Poor-quality images, including blurred images, and those with mucus or foam that prevented adequate mucosal inspection were excluded from the study. The personal data of the patients in the images were hidden. This study was approved by the ethics committee of Shanghai Changhai Hospital (No. CHEC2019-002).
All superficial ESCC and HGIN images were obtained from patients who underwent subsequent endoscopic submucosal dissection. Additionally, histologic results including margin status and invasion depth of endoscopic submucosal dissection specimens were retrieved. Three experienced gastro
All images were annotated by the three endoscopists mentioned above. The first endoscopist reviewed and marked the WLI and NBI images (margins of the lesion) using specific frames. The second endoscopist reviewed the annotations of the first, and if in agreement, the annotation was completed. Alternately, the images were reviewed by a third endoscopist, and the final decision was made in unison. Paris classification subtypes of lesions were also recorded[14].
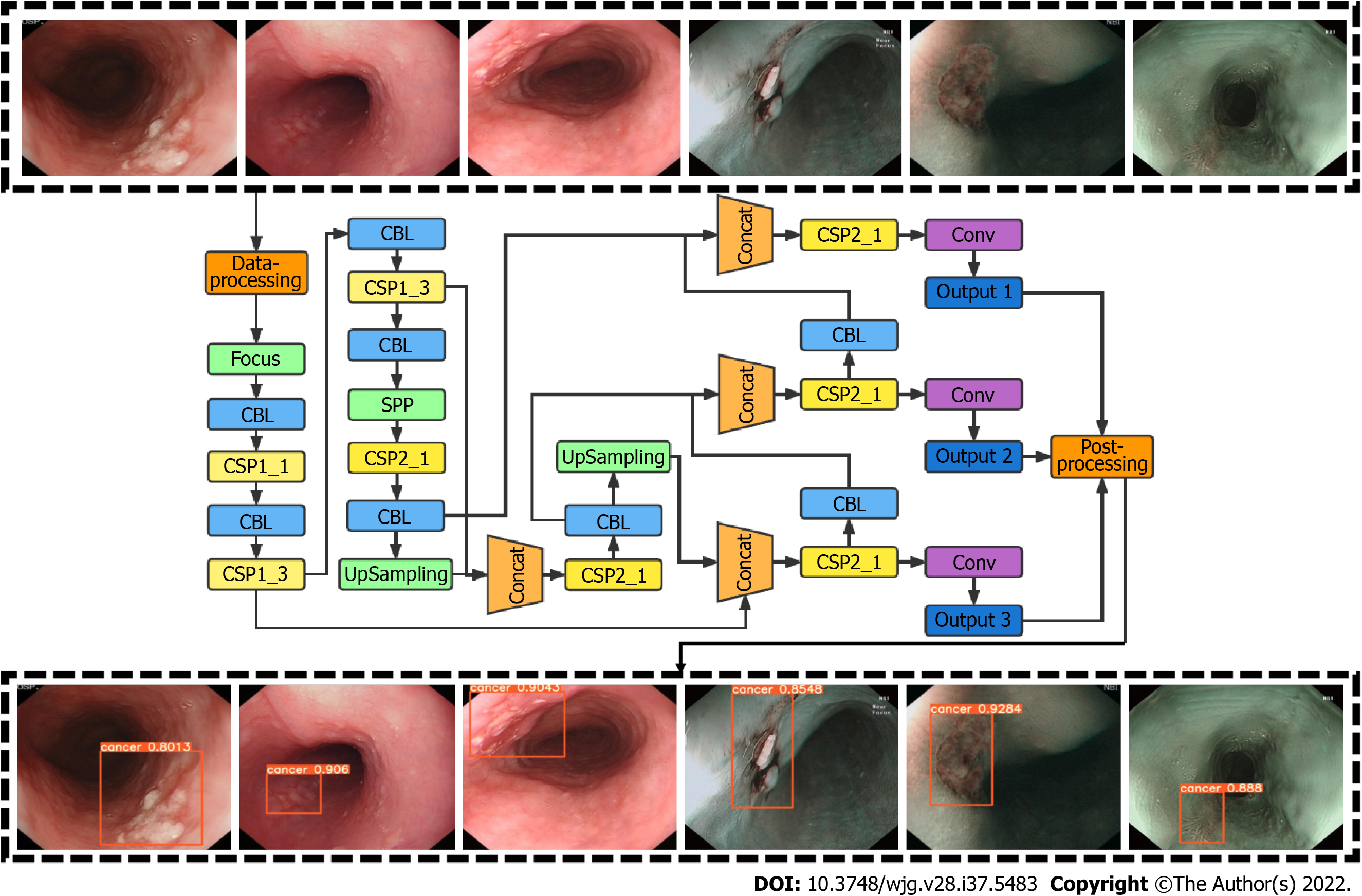
CAD system development was based on the YOLO v5 deep-learning algorithm. YOLO v5 employs cross stage partial network as the backbone for feature extraction and path aggregation network to generate a feature pyramid network to perform feature aggregation and pass it to the prediction mode. YOLO v5 demonstrated increased efficiency and efficacy compared to previous versions. To improve network accuracy, we modified the loss function of YOLO V5 and introduced the mapping distance of the aspect ratio. The input of this network was esophageal images, and the output was an indicator frame that captured the rectangular region of interest, together with the predicted quantitative level of confidence for ESCC above the frame (Figure 1).
The diagnostic accuracy of the CAD system was validated using a previously established independent testing dataset. To compare the performance of the CAD system with that of endoscopists and investigate its added value, four endoscopists from Changhai Hospital were invited to review the images in the testing dataset, independently and with the assistance from the CAD system. Two experts (endoscopy experience ≥ 10 years) and two non-expert endoscopists (endoscopy experience > 1 year but < 5 years) participated. None were involved in the selection and annotation of images. All images in the testing dataset, including 598 WLI and 817 NBI cancerous and 209 WLI and 116 NBI non-cancerous images, were randomly sequenced, and all participating endoscopists independently reviewed the images and made diagnoses within one day. After a washout period of one month, the images were randomly re-sequenced, and endoscopists reviewed the images using the results of the CAD system as a reference. Throughout the study, participating endoscopists were unaware of the correct endoscopic and histologic diagnoses, their own performance, and the diagnostic accuracy of the CAD system.
Primary outcome measures included the area under the receiver operating characteristic curve (AUROC), accuracy, sensitivity, and specificity of the CAD system. Secondary outcome measures included positive predictive value (PPV) and negative predictive value (NPV) of the CAD system; diagnostic performance of participating endoscopists; and improvement in diagnostic accuracy, sensitivity, and specificity in the expert and non-expert groups, after referring to the CAD system results.
AUROC, accuracy, sensitivity, specificity, PPV, and NPV were calculated, and the binominal 95% confidence intervals (CI) were estimated using the CAD-predicted confidence ≥ 0.5, as the prespecified criteria for testing positive. The Delong test was used to compare different AUROCs. A two-sided McNemar test was used to compare differences in accuracy, sensitivity, and specificity. Differences in more than three groups were compared using Cochran’s Q test. Additionally, the McNemar test was used for post-hoc comparisons of P values adjusted using Bonferroni’s method. Pearson’s chi-squared test, t test, and Mann-Whitney U test were used wherever applicable. Superficial ESCC lesions with a CAD-predicted confidence < 0.5 and an intersection over union (IoU) value < 0.45 were defined as inaccurately identified. A P value < 0.05 was considered statistically significant. All statistical analyses were performed using SPSS 26.0 (SPSS Inc., Chicago, IL, United States). The statistical methods of this study were reviewed by XFY from Department of Health Statistics, Naval Medical University.
Table 1 summarizes the characteristics of the patients and lesions in the testing dataset. For 209 cases of superficial ESCC, the lesion size was 2.0 (1.2-4.0) cm and most (82.3%) were located in the middle thoracic esophagus. Flat-type (0-IIb) lesions accounted for 53.6%, and most lesions involved < 25% of the esophageal circumference. Regarding histologic depth of invasion, 165 Lesions (78.9%) were limited to the lamina propria (LPM), 38 (18.2%) invaded the muscularis mucosa or superficial submucosa, and 6 (2.9%) extended beyond the superficial submucosa. Of the 114 non-cancerous cases, 23 were diagnosed with gastroesophageal reflux disease, five were diagnosed with esophageal submucosal lesions, and 86 were judged to be normal on upper gastrointestinal endoscopy.
| Characteristics | Values |
| Cancerous cases (n = 209) | |
| Age (yr), mean ± SD | 62.0 ± 7.2 |
| Sex, Male/Female | 158/51 |
| Lesion location, Ce/Ut/Mt/Lt/Ae | 2/21/172/14/0 |
| Lesion size (cm), median (Q1, Q3) | 2.0 (1.2-4.0) |
| Paris classification, 0-I/IIa/IIb/IIc/IIa+IIc/III | 6/43/112/26/20/2 |
| Circumference, < 1/4, 1/4-1/2, 1/2-3/4, > 3/4 | 117/63/18/11 |
| Depth of invasion, EP-LPM/MM-SM1/SM2 | 165/38/6 |
| Non-Cancerous cases (n = 114) | |
| Age (yr), mean ± SD | 63.2 ± 5.3 |
| Sex, male/female | 65/49 |
| Endoscopic diagnosis, GERD/submucosal lesion/normal | 23/5/86 |
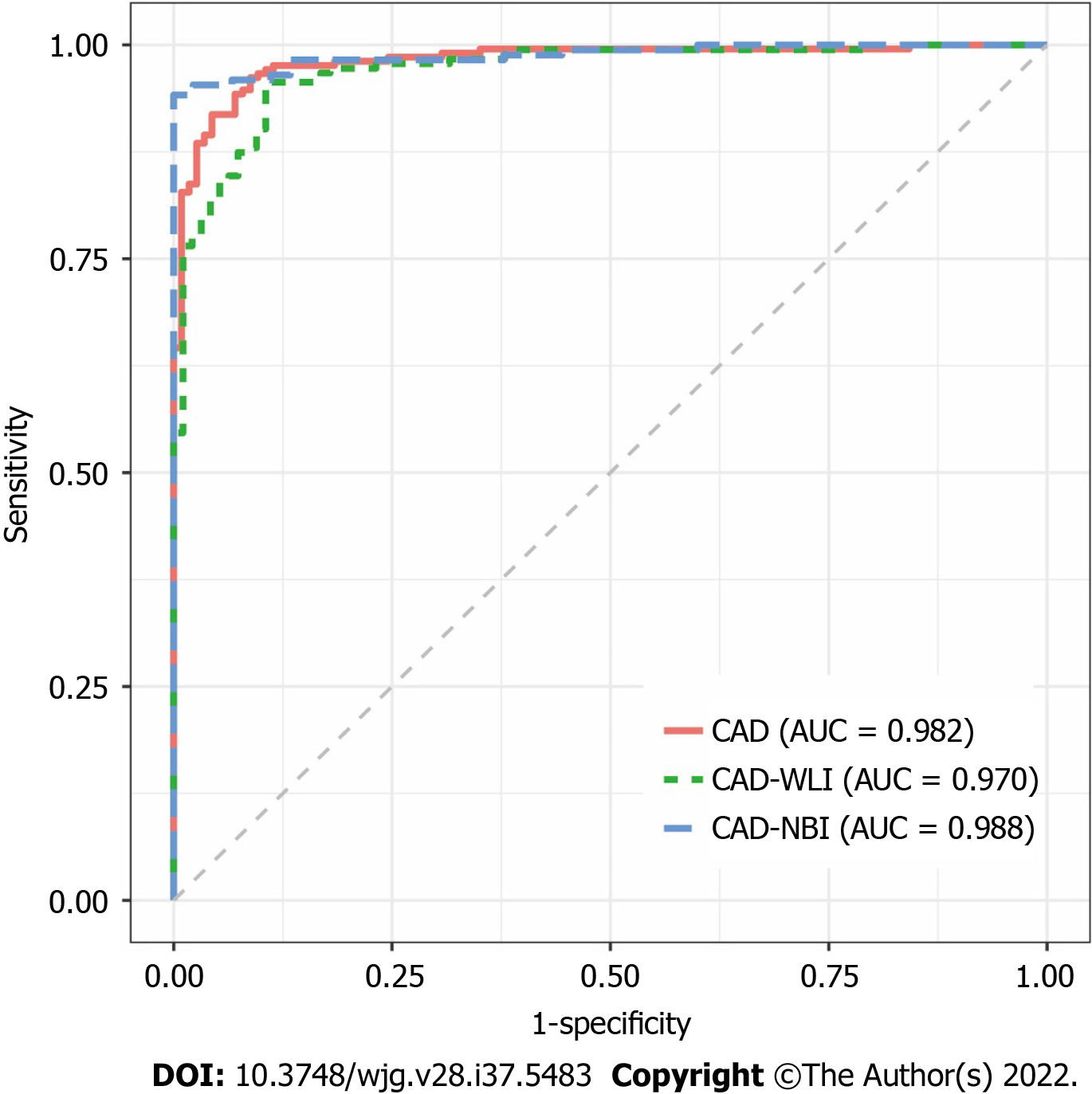
The receiver operating characteristic (ROC) curve of the CAD system is shown in Figure 2. The AUROC of the WLI, NBI, and combined modes were 0.970, 0.988, and 0.982, respectively. The AUROC of WLI, NBI, and the combined mode did not differ significantly. The diagnostic performance measures of the CAD system in the testing dataset are summarized in Table 2. In per-patient analysis, the accuracy, sensitivity, specificity, PPV, and NPV of the CAD system were 92.9%, 91.9%, 94.7%, 97.0%, and 86.4%, respectively. The accuracy (P < 0.001), sensitivity (P < 0.001), and specificity (P = 0.015) of the CAD system were significantly different among the different imaging modes. In post-hoc comparisons, the accuracy of the NBI (adjusted P < 0.001) and combined modes (adjusted P = 0.002) was significantly higher than that of the WLI mode. Furthermore, the sensitivity of NBI (adjusted P < 0.001) and combined modes (adjusted P = 0.014) was significantly higher than that of the WLI mode. No statistically significant differences in specificity were found in post-hoc comparisons between the two imaging modes. The results of the per-imaging analysis are presented in Table 2.
| Per-patient analysis | Per-image analysis | |||||||
| Overall (%) | WLI (%) | NBI (%) | P value | Overall (%) | WLI (%) | NBI (%) | P value | |
| Accuracy (95%CI) | 92.9 (89.5-95.2) | 89.2 (85.3-92.1) | 94.7 (91.7-96.7) | < 0.001 | 87.5 (85.9-89.0) | 85.9 (83.3-88.1) | 89.0 (86.8-90.9) | 0.156 |
| Sensitivity (95%CI) | 91.9 (87.4-94.9) | 88.0 (82.9-91.8) | 94.3 (90.2-96.7) | < 0.001 | 86.6 (84.7-88.3) | 84.1 (80.8-86.8) | 88.4 (86.0-90.4) | 0.067 |
| Specificity (95%CI) | 94.7 (89.0-97.6) | 91.2 (84.6-95.2) | 95.6 (90.1-98.1) | 0.015 | 91.7 (88.2-94.3) | 90.9 (86.2-94.1) | 93.4 (87.0-96.8) | 0.750 |
| PPV (95%CI) | 97.0 (93.5-98.6) | 94.8 (90.8-97.2) | 97.7 (94.3-98.9) | 0.316 | 97.9 (96.9-98.5) | 96.2 (94.2-97.6) | 99.0 (98.0-99.5) | 0.004 |
| NPV (95%CI) | 86.4 (79.3-91.3) | 80.6 (73.0-86.5) | 90.1 (83.5-94.2) | 0.099 | 61.1 (56.6-65.4) | 67.4 (61.7-72.6) | 51.8 (44.8-58.8) | 0.003 |
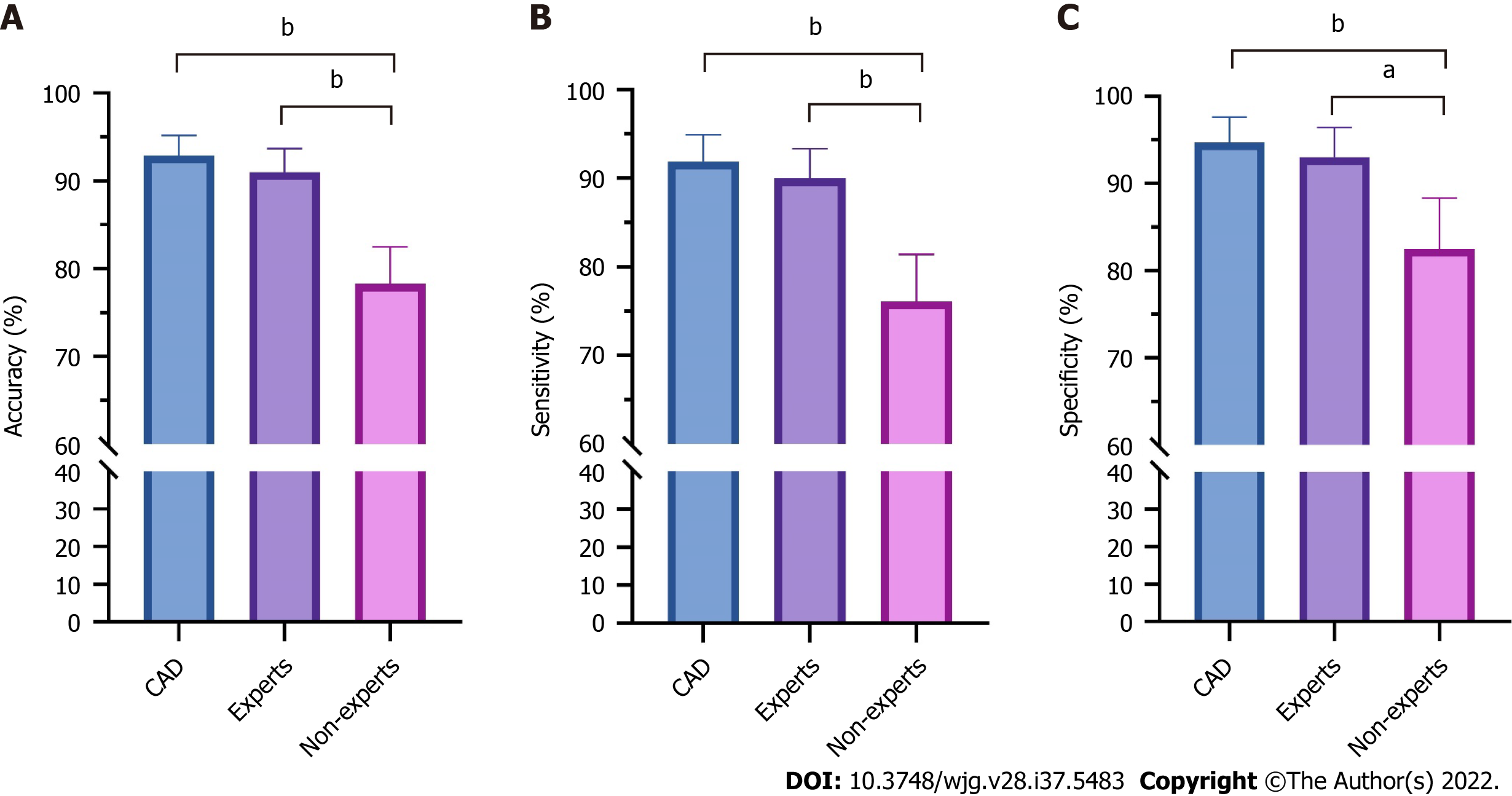
Figure 3 compares the diagnostic performance of the CAD system under the combined WLI and NBI modes, among endoscopists with different experience levels. The accuracy (91.0% vs 78.3%, P < 0.001), sensitivity (90.0% vs 76.1%, P < 0.001), and specificity (93.0% vs 82.5%, P = 0.002) were significantly higher for expert endoscopists than for non-expert endoscopists. The diagnostic performance of the CAD system was significantly higher than that of non-expert endoscopists (P < 0.001) and similar to that of expert endoscopists.
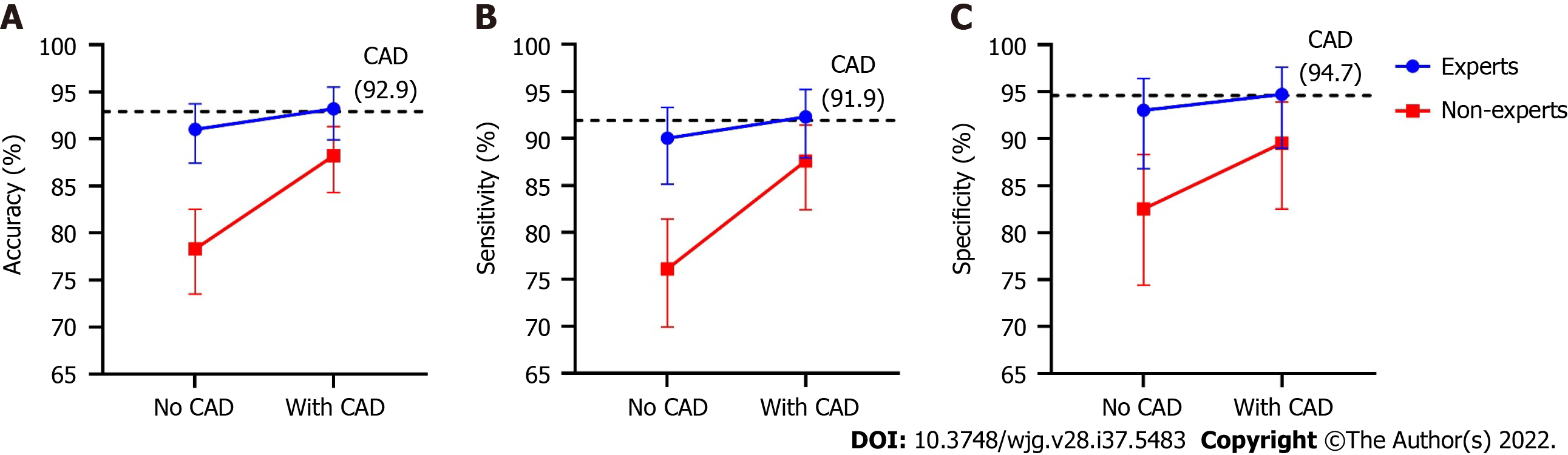
Figure 4 depicts improved diagnostic performance of endoscopists for superficial ESCC after referring to the results of the CAD system. The accuracy (88.2% vs 78.3%, P < 0.001), sensitivity (87.6% vs 76.1%, P < 0.001), and specificity (89.5% vs 82.5%, P = 0.039) of the non-expert endoscopists were all significantly improved after using the CAD system, whereas the diagnostic performances of expert endoscopists did not differ significantly between the two stages. After referring to the CAD system, the accuracy (88.2% vs 93.2%, P < 0.001) and sensitivity (87.6% vs 92.3%, P = 0.013) of the non-experts were still significantly lower than those of the experts, while the disparities were markedly decreased. The specificity (89.5% vs 94.7%, P = 0.124) was similar between experts and non-experts after referring to the CAD.
Under the prespecified definition, 23 (11.0%) lesions were inaccurately identified. Table 3 compares the characteristics of lesions with accurate and inaccurate identifications. The proportion of type 0-IIb lesions was significantly higher in inaccurately identified lesions (73.9% vs 51.1%, P = 0.038), and the proportion of lesions limited to the LPM was higher in the inaccurate identification group, although not significantly so (91.3% vs 77.4%, P = 0.123).
| Characteristics | Accurate identification (n = 186) | Inaccurate identification (n = 23) | P value |
| Lesion location, n (%) | 1.000 | ||
| Ce | 2 (1.1) | 0 (0) | |
| Ut | 13 (7.0) | 2 (8.7) | |
| Mt | 152 (81.7) | 20 (87.0) | |
| Lt | 19 (10.2) | 1 (4.3) | |
| Lesion size (cm), median (Q1, Q3) | 2.0 (1.1-4.0) | 3 (1.5-3.0) | 0.476 |
| Paris classification, n (%) | 0.038 | ||
| Type 0-IIb | 95 (51.1) | 17 (73.9) | |
| Other non-flat types | 91 (48.9) | 6 (26.1) | |
| Circumference, n (%) | 0.591 | ||
| < 1/4 | 107 (57.4) | 10 (43.5) | |
| 1/4-1/2 | 54 (29.0) | 9 (39.1) | |
| 1/2-3/4 | 16 (8.6) | 2 (8.7) | |
| > 3/4 | 9 (4.8) | 2 (8.7) | |
| Depth of invasion, n (%) | 0.123 | ||
| EP-LPM | 144 (77.4) | 21 (91.3) | |
| MM and deeper | 42 (22.6) | 2 (8.7) |
Upper gastrointestinal endoscopy is critical for screening and early diagnosis of ESCC. However, the subtle endoscopic appearance of most superficial ESCC types is challenging for early detection and relies greatly on the skills and experience of endoscopists. Herein, we developed and validated a novel deep-learning-based CAD system for the detection of superficial ESCC in the WLI and NBI modes of upper gastrointestinal endoscopy. The AUROC, accuracy, sensitivity, and specificity of the CAD system were 0.982, 92.9%, 91.9%, and 94.7%, respectively. The diagnostic performance was significantly superior to that of non-experts and comparable to that of expert endoscopists. After referring to the results of the CAD system, the detectability of non-expert endoscopists significantly improved, and disparities between experts and non-experts markedly decreased. This system may assist in improving the detectability of superficial ESCC, particularly for inexperienced endoscopists and in undeveloped areas with limited resources.
Horie et al[9] developed a deep learning-based CAD system for superficial ESCC, with a sensitivity of 98% and low PPV of 40%. Zhao et al[15] developed an AI-based model for magnifying NBI images for automated classification of intrapapillary capillary loops, with a mean diagnostic accuracy of 89.6%. In a recent study by Guo et al[16], the authors developed a CAD system for real-time automated diagnosis of precancerous lesions and ESCC based on endoscopic video datasets, which achieved a per-lesion sensitivity of 100%. A systematic review by Visaggi et al[13] reported that the AUROC, sensitivity, and specificity of AI in diagnosing ESCC were 0.97, 0.95, and 0.92, respectively. However, few existing systems can support WLI and NBI, both of which are indispensable to the diagnosis of superficial ESCC under the same algorithm framework. We developed the current system under the updated YOLO v5 algorithm network supporting WLI and NBI, and achieved an AUROC, accuracy, sensitivity, and specificity of 0.982, 92.9%, 91.9%, and 94.7%, respectively, which was comparable to the best per
Superficial ESCC is difficult to inspect under WLI; however, NBI may significantly improve detectability with similar sensitivity and higher specificity than Lugol chromoendoscopy[6]. This may explain the demonstrated decrease in the accuracy and sensitivity of the CAD system, without NBI. Cai et al[10] reported on a deep neural network (DNN)-based system that utilizes WLI with high accuracy and sensitivity, for early ESCC detection. They subsequently developed a CAD-NBI system and found that its accuracy (94.3% vs 89.5%, P = 0.028) and specificity (96.7% vs 83.1%, P < 0.001) were significantly better than those of the CAD-WLI system[12]. By using CAD-WLI and CAD-NBI, endoscopists can significantly improve their diagnostic efficacy. Taken together, we recommend the combined implementation of NBI with other image-enhancing techniques for the detection of superficial ESCC, even under the CAD system.
To assess the clinical application value of the CAD system, endoscopists of varying experience reviewed the entire testing dataset independently and after referring to the diagnostic results of the CAD system. During the independent review, non-expert endoscopists showed significantly lower accuracy, sensitivity, and specificity than the experts, which highlighted the difficulty of novices in detecting superficial ESCC and disparities in detectability among endoscopists. The diagnostic performance of the non-experts significantly improved after referring to the CAD system. This suggests that the results of the CAD system provide useful references for non-expert endoscopists and facilitate further inspection and evaluation of potentially malignant lesions. The improved diagnostic performance of non-experts remained unequal to that of the CAD system, suggesting that endoscopists had made independent decisions, rather than relying completely on CAD. Similar results were reported in previous studies[10,12,17]. In the current study, although the performance of non-experts was still inferior to that of the experts after referring to the CAD system, the disparity markedly decreased. This suggests that the CAD system can homogenize the performance of endoscopists by overcoming the differences among endoscopists with different experience levels. In clinical practice, the usefulness of high-definition and imaging-enhanced endoscopy may be jeopardized by the inadequate number of highly trained endoscopists. In contrast, the CAD system does not require additional training and has instead been found to improve the performance of non-expert endoscopists by approximating the level of expertise. Thus, the CAD system may be especially useful in resource-limited areas, to improve the quality of endoscopic services and patient outcomes.
To our knowledge, this is the first study to analyze the characteristics of inaccurately identified lesions. We defined cases of inaccurate identification based on the predicted confidence and accuracy of the location frame. Endoscopists should be aware that flat-type and earlier-stage lesions limited to the LPM may be associated with an increased risk of inaccurate identification. To avoid a missed diagnosis of superficial ESCC, we recommend the use of Lugol’s staining or magnifying endoscopy for flat-type lesions that are difficult to distinguish even under CAD.
In clinical practice, the CAD system automatically captures images as endoscopists inspect the esophagus, once integrated into an endoscopy workstation. Additionally, images frozen by endosco
Our study had several limitations. First, the number of included endoscopists was relatively small, and the representation might be inadequate. Second, only still and non-magnified images were included; thus, the identification of videos and types of intrapapillary capillary loops could not be achieved. Third, the numbers and angles of lesions in NBI and WLI images were not identical in the testing dataset, and biases may exist in the comparison of diagnostic performance among the different modes of the CAD system.
We developed and validated a deep-learning CAD system, for the endoscopic detection of superficial ESCC, that achieved a high diagnostic accuracy comparable to that of expert endoscopists and could significantly improve the detecting ability of non-expert endoscopists.
Esophageal squamous cell carcinoma (ESCC) is a leading cause of cancer-related morbidity and mortality worldwide. Upper gastrointestinal endoscopy is critical for ESCC detection; however, endoscopists require long-term training to avoid missing superficial lesions. Artificial intelligence (AI) has been increasingly investigated to assist endoscopic diagnosis.
AI has shown promising results for endoscopic diagnosis of superficial ESCC. However, few AI-based computer-assisted diagnosis (CAD) systems for ESCC that support white-light and narrow-band imaging have been applied in clinical practice.
We aimed to develop a CAD system for endoscopic detection of superficial ESCC and investigate its application value.
We configured the CAD system for white-light and narrow-band imaging modes based on the YOLO v5 algorithm. A total of 4447 images from 837 patients and 1695 images from 323 patients were included in the training and testing datasets, respectively. Two experts and two non-expert endoscopists reviewed the testing dataset independently and with computer assistance. The diagnostic performance was evaluated in terms of the area under the receiver operating characteristic curve, accuracy, sensitivity, and specificity.
The area under the receiver operating characteristics curve, accuracy, sensitivity, and specificity of the CAD system were 0.982 [95% confidence interval (CI): 0.969-0.994], 92.9% (95%CI: 89.5%-95.2%), 91.9% (95%CI: 87.4%-94.9%), and 94.7% (95%CI: 89.0%-97.6%), respectively. The accuracy of CAD was significantly higher than that of non-expert endoscopists (78.3%, P < 0.001 compared with CAD) and comparable to that of expert endoscopists (91.0%, P = 0.129 compared with CAD). After referring to the CAD results, the accuracy of the non-expert endoscopists significantly improved (88.2% vs 78.3%, P < 0.001).
The diagnostic performance of the CAD system is promising and may assist in improving detectability, particularly for inexperienced endoscopists.
An updated CAD system that can process real-time videos or images with suboptimal quality is in development. Randomized controlled trials are warranted to investigate the clinical pragmaticality.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shiroma S, Japan; Sudou K, Japan; Tsoulfas G, Greece S-Editor: Gong ZM L-Editor: A P-Editor: Gong ZM
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 51377] [Article Influence: 8562.8] [Reference Citation Analysis (122)] |
| 2. | Arnold M, Ferlay J, van Berge Henegouwen MI, Soerjomataram I. Global burden of oesophageal and gastric cancer by histology and subsite in 2018. Gut. 2020;69:1564-1571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 284] [Cited by in F6Publishing: 258] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 3. | Chen R, Liu Y, Song G, Li B, Zhao D, Hua Z, Wang X, Li J, Hao C, Zhang L, Liu S, Wang J, Zhou J, Zhang Y, Li Y, Feng X, Li L, Dong Z, Wei W, Wang G. Effectiveness of one-time endoscopic screening programme in prevention of upper gastrointestinal cancer in China: a multicentre population-based cohort study. Gut. 2021;70:251-260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 62] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 4. | Hölscher AH, Bollschweiler E, Schröder W, Metzger R, Gutschow C, Drebber U. Prognostic impact of upper, middle, and lower third mucosal or submucosal infiltration in early esophageal cancer. Ann Surg. 2011;254:802-7; discussion 807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 5. | Lee YC, Wang CP, Chen CC, Chiu HM, Ko JY, Lou PJ, Yang TL, Huang HY, Wu MS, Lin JT, Hsiu-Hsi Chen T, Wang HP. Transnasal endoscopy with narrow-band imaging and Lugol staining to screen patients with head and neck cancer whose condition limits oral intubation with standard endoscope (with video). Gastrointest Endosc. 2009;69:408-417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Morita FH, Bernardo WM, Ide E, Rocha RS, Aquino JC, Minata MK, Yamazaki K, Marques SB, Sakai P, de Moura EG. Narrow band imaging versus lugol chromoendoscopy to diagnose squamous cell carcinoma of the esophagus: a systematic review and meta-analysis. BMC Cancer. 2017;17:54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 7. | Ishihara R, Takeuchi Y, Chatani R, Kidu T, Inoue T, Hanaoka N, Yamamoto S, Higashino K, Uedo N, Iishi H, Tatsuta M, Tomita Y, Ishiguro S. Prospective evaluation of narrow-band imaging endoscopy for screening of esophageal squamous mucosal high-grade neoplasia in experienced and less experienced endoscopists. Dis Esophagus. 2010;23:480-486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 8. | Rodríguez de Santiago E, Hernanz N, Marcos-Prieto HM, De-Jorge-Turrión MÁ, Barreiro-Alonso E, Rodríguez-Escaja C, Jiménez-Jurado A, Sierra-Morales M, Pérez-Valle I, Machado-Volpato N, García-Prada M, Núñez-Gómez L, Castaño-García A, García García de Paredes A, Peñas B, Vázquez-Sequeiros E, Albillos A. Rate of missed oesophageal cancer at routine endoscopy and survival outcomes: A multicentric cohort study. United European Gastroenterol J. 2019;7:189-198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 9. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 220] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 10. | Cai SL, Li B, Tan WM, Niu XJ, Yu HH, Yao LQ, Zhou PH, Yan B, Zhong YS. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2019;90:745-753.e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 11. | Luo H, Xu G, Li C, He L, Luo L, Wang Z, Jing B, Deng Y, Jin Y, Li Y, Li B, Tan W, He C, Seeruttun SR, Wu Q, Huang J, Huang DW, Chen B, Lin SB, Chen QM, Yuan CM, Chen HX, Pu HY, Zhou F, He Y, Xu RH. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645-1654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 209] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 12. | Li B, Cai SL, Tan WM, Li JC, Yalikong A, Feng XS, Yu HH, Lu PX, Feng Z, Yao LQ, Zhou PH, Yan B, Zhong YS. Comparative study on artificial intelligence systems for detecting early esophageal squamous cell carcinoma between narrow-band and white-light imaging. World J Gastroenterol. 2021;27:281-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 13. | Visaggi P, Barberio B, Gregori D, Azzolina D, Martinato M, Hassan C, Sharma P, Savarino E, de Bortoli N. Systematic review with meta-analysis: artificial intelligence in the diagnosis of oesophageal diseases. Aliment Pharmacol Ther. 2022;55:528-540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 14. | Endoscopic Classification Review Group. Update on the paris classification of superficial neoplastic lesions in the digestive tract. Endoscopy. 2005;37:570-578. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 559] [Cited by in F6Publishing: 571] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 15. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 16. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 17. | Yang XX, Li Z, Shao XJ, Ji R, Qu JY, Zheng MQ, Sun YN, Zhou RC, You H, Li LX, Feng J, Yang XY, Li YQ, Zuo XL. Real-time artificial intelligence for endoscopic diagnosis of early esophageal squamous cell cancer (with video). Dig Endosc. 2021;33:1075-1084. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |