Copyright
©The Author(s) 2022.
World J Gastroenterol. Jul 7, 2022; 28(25): 2823-2842
Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2823
Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2823
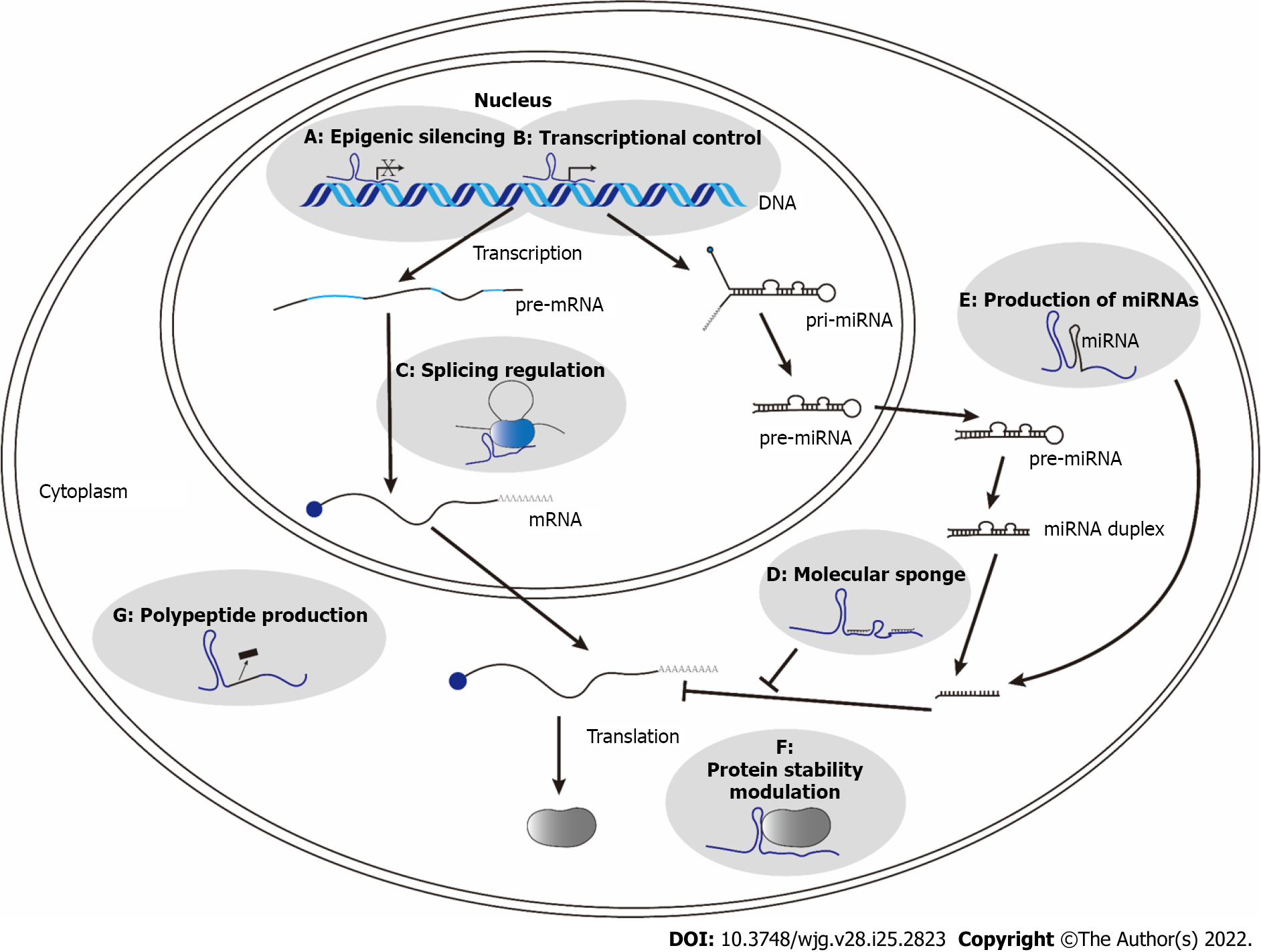
Figure 3 Regulatory mechanisms of various long noncoding RNAs in hepatitis B virus replication and oncogenesis[2].
A: HOX transcript antisense RNA (HOTAIR), high expression in hepatocellular carcinoma, urothelial carcinoma associated 1 (UCA1), HOTAIR, LINC00152, PVT1, and antisense noncoding RNA in the INK4 locus (ANRIL) could regulate gene expression through epigenetic silencing; B: HOXA transcript at the distal tip, LINC00152, and metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) could increase the expression of their target genes through transcriptional control; C: MALAT1 could modulate alternative splicing; D: Unigene56159, highly upregulated in liver cancer (HULC), hepatitis B virus (HBV) X protein-long interspersed nuclear elements 1, UCA1, ANRIL, LINC01149 variant, LINC01352, F11-AS1, LINC01232, n335586, XIST, SNHG5, SSTR5-AS1, and TRERNA1 regulate the gene expression through molecular sponging to sequester miRNAs; E: Long noncoding RNAs (lncRNAs) Ftx could produce miRNAs to regulate their target genes; F: LncRNA-Dreh and HULC could modulate protein stability; G: LncRNA HBVPTPAP could encode a small polypeptide to exert its function.
- Citation: Li HC, Yang CH, Lo SY. Long noncoding RNAs in hepatitis B virus replication and oncogenesis. World J Gastroenterol 2022; 28(25): 2823-2842
- URL: https://www.wjgnet.com/1007-9327/full/v28/i25/2823.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i25.2823