Copyright
©The Author(s) 2022.
World J Gastroenterol. Jul 7, 2022; 28(25): 2782-2801
Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2782
Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2782
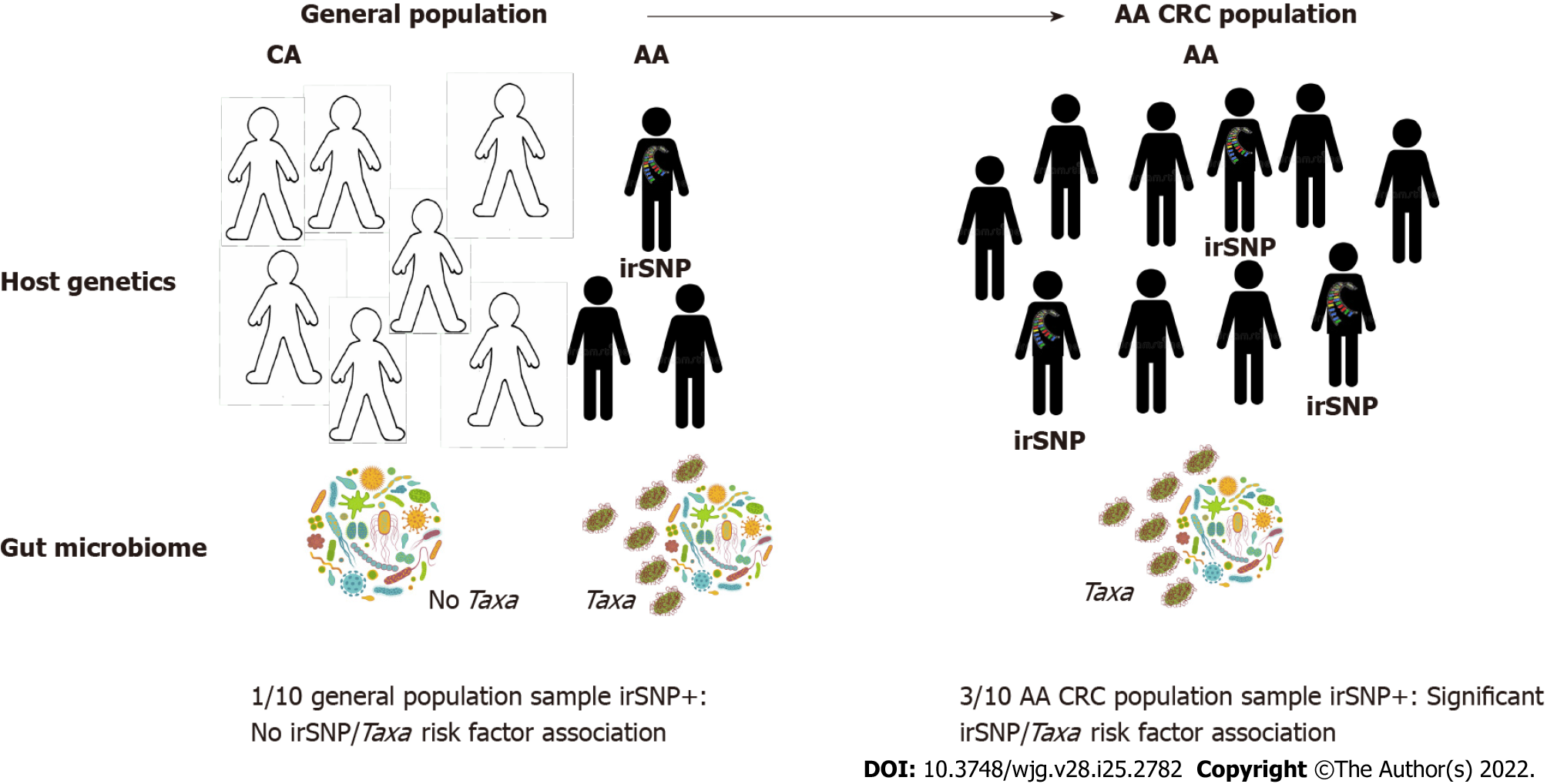
Figure 3 Population-specific colorectal cancer microbiome genome-wide association studies to understand colorectal cancer disparities.
Systemic underrepresentation of minorities in microbiome genome-wide association studies (mGWAS) may compromise the identification of microbiome-associated colorectal cancer (CRC) risks in African-Americans (AAs). Left: Associations between AA-enriched immune-related single-nucleotide polymorphisms (irSNPs) (pop-irSNPs) and gut bacteria (Taxa) may remain undetected in the global population if pop-irSNP and Taxa have too low frequency in the AA population and AAs are underrepresented. Right: If pop-irSNP and pathogenic bacteria are CRC risk factors in AA, they will be enriched in AA CRC patients compared to the general AA population (left), and mGWAS will detect the association between pop-irSNP and Taxa in AA CRC cohort. These features justify population-specific CRC mGWAS to detect additional CRC risk resulting from the interaction between ancestral irSNP and bacteria in minorities. CRC: Colorectal cancer; irSNPs: Immune-related single-nucleotide polymorphisms; mGWAS: Microbiome genome-wide association studies; AA: African-American; CA: Caucasian-American.
- Citation: Ahmad S, Ashktorab H, Brim H, Housseau F. Inflammation, microbiome and colorectal cancer disparity in African-Americans: Are there bugs in the genetics? World J Gastroenterol 2022; 28(25): 2782-2801
- URL: https://www.wjgnet.com/1007-9327/full/v28/i25/2782.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i25.2782