Copyright
©The Author(s) 2021.
World J Gastroenterol. Nov 28, 2021; 27(44): 7687-7704
Published online Nov 28, 2021. doi: 10.3748/wjg.v27.i44.7687
Published online Nov 28, 2021. doi: 10.3748/wjg.v27.i44.7687
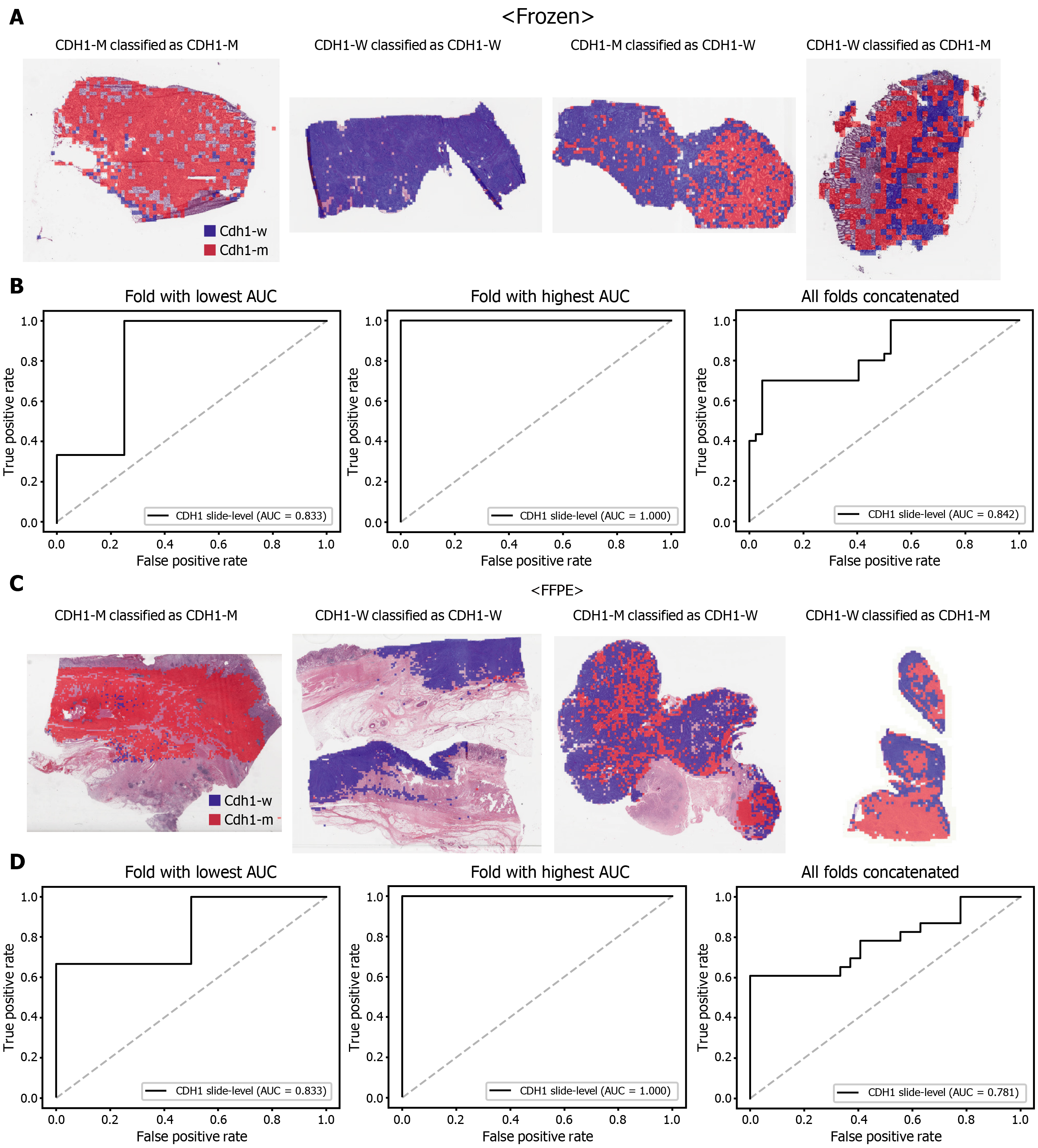
Figure 2 Classification results of CDH1 gene in the The Cancer Genome Atlas gastric cancer dataset.
A: Representative whole slide images (WSIs) of the frozen slides with CDH1 gene mutation correctly classified as mutation, with wild-type gene correctly classified as wild-type, with gene mutation falsely classified as wild-type, and with wild-type gene falsely classified as mutation, from left to right; B: Receiver operating characteristic (ROC) curves for the fold with lowest area under the curve (AUC), for the fold with highest AUC, and for the concatenated results of all ten folds, from left to right, obtained with the classifiers trained with the frozen tissues; C and D: Same as A and B but the results were for the formalin-fixed paraffin-embedded WSIs. CDH1-M: CDH1 mutated, CDH1-W: CDH1 wild-type.
- Citation: Jang HJ, Lee A, Kang J, Song IH, Lee SH. Prediction of genetic alterations from gastric cancer histopathology images using a fully automated deep learning approach. World J Gastroenterol 2021; 27(44): 7687-7704
- URL: https://www.wjgnet.com/1007-9327/full/v27/i44/7687.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i44.7687