Published online Oct 14, 2021. doi: 10.3748/wjg.v27.i38.6387
Peer-review started: January 14, 2021
First decision: March 29, 2021
Revised: April 30, 2021
Accepted: September 6, 2021
Article in press: September 6, 2021
Published online: October 14, 2021
Specificity protein (Sp) transcription factors (TFs) Sp1, Sp3 and Sp4, and the orphan nuclear receptor 4A1 (NR4A1) are highly expressed in pancreatic tumors and Sp1 is a negative prognostic factor for pancreatic cancer patient survival. Results of knockdown and overexpression of Sp1, Sp3 and Sp4 in pancreatic and other cancer lines show that these TFs are individually pro-oncogenic factors and loss of one Sp TF is not compensated by other members. NR4A1 is also a pro-oncogenic factor and both NR4A1 and Sp TFs exhibit similar functions in pancreatic cancer cells and regulate cell growth, survival, migration and invasion. There is also evidence that Sp TFs and NR4A1 regulate some of the same genes including survivin, epidermal growth factor receptor, PAX3-FOXO1, α5- and α6-integrins, β1-, β3- and β4-integrins; this is due to NR4A1 acting as a cofactor and mediating NR4A1/Sp1/4-regulated gene expression through GC-rich gene promoter sites. Several studies show that drugs targeting Sp downregulation or NR4A1 antagonists are highly effective inhibitors of Sp/NR4A1-regulated pathways and genes in pancreatic and other cancer cells, and the triterpenoid celastrol is a novel dual-acting agent that targets both Sp TFs and NR4A1.
Core Tip: Specificity protein (Sp), transcription factors (TFs), Sp1, Sp3 and Sp4, and nuclear receptor 4A1 (NR4A1, Nur77) are highly expressed in pancreatic cancer cells and tumors. Results of gene silencing studies show that Sp TFs and NR4A1 are pro-oncogenic and regulate pathways/genes associated with cell proliferation, survival and migration/invasion. Bis-indole derived ligands (CDIMs) that bind NR4A1 act as NR4A1 antagonists and we discuss an important mechanism of gene regulation by NR4A1/Sp complexes which can be inhibited by NR4A1 antagonists.
- Citation: Safe S, Shrestha R, Mohankumar K, Howard M, Hedrick E, Abdelrahim M. Transcription factors specificity protein and nuclear receptor 4A1 in pancreatic cancer. World J Gastroenterol 2021; 27(38): 6387-6398
- URL: https://www.wjgnet.com/1007-9327/full/v27/i38/6387.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i38.6387
Pancreatic cancer is a devastating disease and it is estimated that in 2020, the number of diagnosed cases in the United States will be 57600 and 47050 will die because of this disease[1]. Despite advances in treating pancreatic cancer with surgical intervention and various therapeutic regimens, the five-year survival rate from pancreatic cancer is < 10%[1] and this is due, in part to the late detection of the tumors due to lack of symptoms and lack of early biomarkers of this cancer. There are many risk factors for pancreatic cancer and this includes pancreatitis, obesity and metabolic syndrome; due to the obesity crisis in many countries, it is estimated that by 2030, pancreatic cancer will be the second leading cause of cancer deaths[2]. Development of pancreatic cancer is associated with the temporal increases in expression of various oncogenes and inactivation of tumor suppressor genes resulting in enhanced cancer cell proliferation, survival, migration, invasion and metastasis[3-6]. Gemcitabine has largely replaced 5-fluorouracil as the major cytotoxic drug for treatment of pancreatic cancer and many other mechanism-based drugs targeting critical genes associated with various pro-oncogenic pathways in pancreatic cancer are being developed or are in clinical trials[7,8]. The success of these agents in combination therapies targeting one or more pro-oncogenic pathways has been limited but promising. For example, one study using the vascular endothelial growth factor inhibitor bevacizumab in combination with leucovorin, nab-paclitaxel, oxiplatin and 5-fluorouracil and this resulted in a one-year survival of 82% for the treated patients[9]. In this review, we will focus on three transcription factors (TFs), namely specificity proteins (Sps) Sp1, Sp3 and Sp4, and nuclear receptor 4A1 (NR4A1, TR3, Nur77) and their overlapping pro-oncogenic roles in pancreatic cancers. We will also highlight an interesting convergence of these TFs and outline approaches for simultaneous targeting NR4A1 and Sp TFs by mechanism-based agents resulting in inhibition of cancer cell growth, survival, migration and invasion.
Sp TFs are members of the Sp-Kruppel-like factor family (Sp/KLF) of TFs which includes 9 Sp TFs and 17 KLF proteins[10-14]. Sp1-Sp4 genes have a similar domain structure, however, it is evident that among other Sp/KLF family of TFs, their domain structures are highly diverse with the most common features being the zinc finger DNA binding domain and its subsequent interactions with GC/GT-rich gene promoter sequences[10-14]. Sp1 was the first TF identified and characterized[15] and Sp1 knockout mouse embryos exhibited retarded development and other abnormalities, and embryolethality is observed (day 11)[11]. Comparable studies on other Sp family members indicate that these TFs play important roles in early development and differentiation by their regulation of key genes involved in these functions. A detailed analysis of the age-dependent expression of individual Sp TFs is not available, however, there is evidence that for Sp1, there is an age-dependent decrease in expression in both rodent and human tissues[16-19]. Presumably the functions of Sp1 in aging animals are replaced by other TFs. The concern and subsequent focus on Sp1 and other Sp TFs (Sp3 and Sp4) in cancer emerged from studies showing that Sp1, Sp3 and Sp4 were overexpressed in cancer cells and tumors[20]. These TFs were also identified as negative prognostic factors for cancer patient survival from several cancers and functional studies demonstrated the pro-oncogenic activity for Sp TFs[20]. The overexpression and prognostic value of Sp TFs have primarily focused on Sp1 and one of the first studies showed that Sp1 was overexpressed in patients with pancreatic tumors and was a negative prognostic factor for patient survival[21]. Most studies on cancer patients have reported that Sp1 or Sp3 is overexpressed and/or is a negative prognostic factor for glioma, astrocytoma, colon, gastric, liver, prostate and head and neck cancers; in lung and breast cancers; in addition, there are some conflicting results[22-40]. An important linkage between Sp1 and cancer was observed in studies showing that carcinogen- or oncogene- induced transformation of human fibroblasts into fibrosarcomas was accompanied by an 8-18-fold increase in expression of Sp1. Moreover, the ability of fibrosarcoma cells to form tumors in athymic nude mice was abrogated after Sp1 knockdown[41]. Subsequent studies show that Sp TFs are pro-oncogenic and regulate pathways and genes associated with cell growth, survival, migration and invasion (Figure 1).
NR4A1 and two related receptors NR4A2 (Nurr1) and NR4A3 (Nor1) are orphan nuclear receptors with structures and endogenous functions that differ significantly from Sp TFs[42,43]. NR4A1 and NR4A3 knockout mice are viable whereas NR4A2-/- mice exhibit early mortality due to dopaminergic and other neuronal deficits[44-48]. Endogenous ligands for NR4A have not been identified, however, NR4A bind structurally diverse synthetic compounds and these receptors interact with nerve growth factor β response elements (NBREs: AAAGGTCA) and Nur response elements [NuRE: AAAT(GA)C/T/CA] as monomers and dimers respectively[49-51]. In addition, NR4A1 and NR4A2 form a heterodimer with retinoid X receptor and bind a DR5 motif[52,53]. NR4As are immediate early genes induced by diverse stressors and they play important roles in maintaining cellular homeostasis and in pathophysiology[42,43,54]. NR4A1 and Sp TFs are remarkably distinct in their structures and functions in maintaining cellular homeostasis, however, in pancreatic cancer and other tumor types, their functions are similar. Like Sp1, NR4A1 is overexpressed in solid tumors from patients with pancreatic, breast, liver, glioblastoma, ovarian, colon, melanoma, endometrial, cervical, rhabdomyosarcoma and gastric cancer[55-64]. Moreover, high expression of NR4A1 in tumors is a negative prognostic factor for lung, breast, colon and ovarian cancer patients[58-60,63]. The negative prognostic significance of NR4A1 overexpression is paralleled by studies showing that like Sp TFs, NR4A1 is pro-oncogenic and regulates pathways comparable to that described for Sp TFs (Figure 1). Thus, there is an interesting parallel in the expression, functions and prognostic values of Sp TFs (primarily Sp1) and NR4A1 in multiple tumor types. This review will focus on the individual roles and interactions of Sp TFs and NR4A1 in pancreatic cancer and this will include genes commonly regulated by NR4A1/Sp complex in which NR4A1/Sp1 and NR4A1/Sp4 interact at GC-rich gene promoter sites that bind Sp.
Shi et al[65] first reported the overexpression of Sp1 in pancreatic cancer cell lines and in pancreatic tumors compared to non-tumor tissue and subsequent studies showed that Sp1, Sp3 and Sp4 were co-expressed in most pancreatic cancer cells[66]. The three structurally related TFs target similar GC-rich sequences in gene promoters and there is evidence for some genes, that Sp3 acts as a transcriptional repressor. A systematic study of the functions of Sp1, Sp3 and Sp4 was investigated in Panc1, MiaPaCa2 and L3.6pL pancreatic cancer cells and also in cell lines derived from lung, colon, kidney and breast tumors[66]. Figure 1 illustrates the functional effects of Sp1 knockdown which resulted in decreased pancreatic cancer cell growth, induced Annexin V staining (apoptosis) and decreased migration and this was accompanied by PARP cleavage, another marker of apoptosis. These results were somewhat surprising since it might be expected that the loss of Sp1 would be compensated for by Sp3 and Sp4. The functional properties of Sp3 and Sp4 were also determined in Panc1, L3.6pL and MiaPaca2 cells by knockdown and the results showed that like Sp1, knockdown of Sp3 or Sp4 decreased cell proliferation, induced apoptosis (increased Annexin V staining and cleaved PARP) and decreased cell invasion and similar results were observed in lung, breast, kidney and colon cancer cell lines[66]. These results indicated the importance of all three Sp TFs as independent pro-oncogenic factors that regulate pancreatic cell growth, survival and invasion, and related genes and it was suggested that Sp TFs were non-oncogene addiction genes[66].
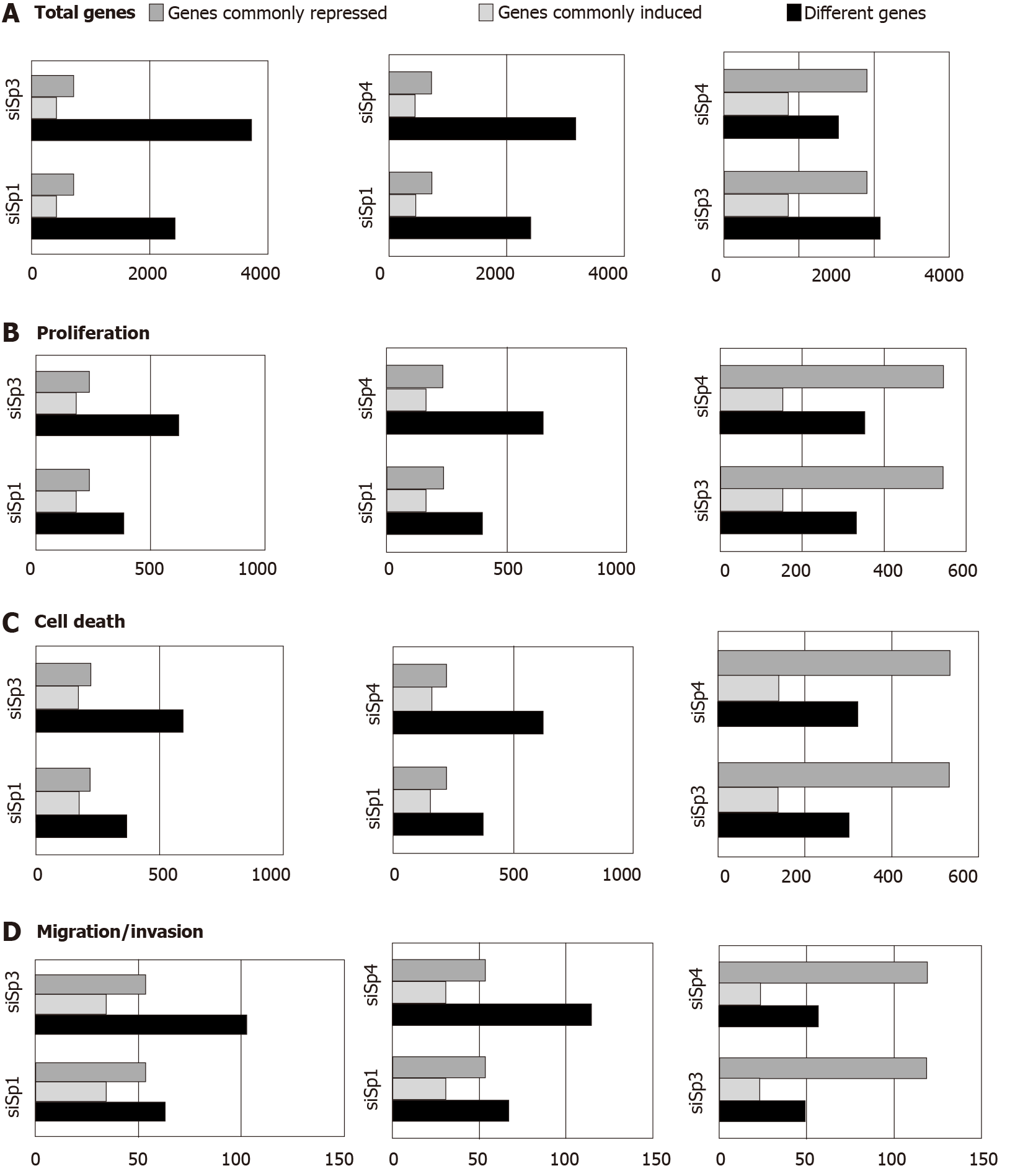
Genomic analysis of altered gene expression after individual knockdown of Sp1, Sp3 and Sp4 in Panc1 pancreatic cancer cells further demonstrated their pro-oncogenic functions and unique effects of Sp1, Sp3 and Sp4 in this cell line. Figure 2A illustrates the total number of genes that are changed after individual knockdown of Sp1, Sp3 and Sp4, the number of uniquely modified genes and the number of genes commonly repressed or induced by Sp1-Sp3, Sp1-Sp4 and Sp3-Sp4. Knockdown of Sp3 and Sp4 resulted in the highest number (and percentage) of commonly induced or repressed genes. Causal Ingenuity Pathway Analysis (IPA) of the genes affected by knockdown were sorted into functional groups and Figure 2B-D illustrate the overlap in affected genes in the Sp1-Sp3, Sp1-Sp4 and Sp3-Sp4 groups associated cell growth, survival and cellular movement (migration and invasion). The overall number and percentage of commonly induced or repressed genes was highest for Sp3 and Sp4 in each functional category and this was similar to that observed for the total number of commonly induced or repressed genes (Figure 2A). Quantitative results from the causal IPA confirmed that the genomic changes observed in the array data were consistent with the functional changes observed after individual knockdown of Sp1, Sp3 and Sp4. This study also confirmed that knockdown of Sp1 alone or Sp1/3/4 (combination) in L3.6pL pancreatic cancer cells also decreased the growth of tumors in athymic nude mice bearing L3.6pL cells as xenografts[66].
In addition to the array studies noted above, there are a number of studies on Sp knockdown or overexpression in pancreatic cancer cells that have identified other Sp-regulated genes that contribute to the pro-oncogenic functions of Sp1, Sp3 and Sp4, and this includes regulation of pro-oncogenic long non-coding RNAs (MALAT1) and microRNAs (Table 1)[20,66-69]. These data were obtained only in pancreatic cancer cells; however, similar results for many of these genes have also been observed in other cancer cell lines[20]. Results summarized in Table 1 were observed in multiple pancreatic cancer cell lines and for some genes, their regulation by Sp TFs is cell context-dependent and there are also differences in the roles of Sp1, Sp3 and Sp4. Some studies used a combination of oligonucleotides targeting the 3 TFs (siSp1/3/4) and the combined knockdown of Sp1, Sp3 and Sp4 by RNA interference in Panc28 and L3.6pL pancreatic cancer cells decreased expression of NFkB-p65 and NFkB-p50 proteins[70]. In the former cell line, individual knockdown of Sp1, Sp3 or Sp4 partially decreased expression of both NFkB subunits. In L3.6pL cells, Sp1 and to a lesser extent Sp3, but not Sp4 knockdown decreased NFkB-p65 whereas Sp4 knockdown was primarily responsible for decreased expression of NFkB-p50 protein.
| Functional | Sp-regulated gene |
| Cell proliferation and cell cycle progression | EGFR, IGFR, cyclin D1, PIK3R1, pmTOR, 4-EBP, pS6RP, STAT3, pSTAT3, NFkB-p65/950 |
| Survival and apoptosis | Survivin, bcl-2, PARP cleavage, APAF1, ATM, TNFRSF25, COX2, NFkB-p65/5 |
| Cell migration/movement EMT | Β1-integrin, α2-integrin, MTSS1, vimentin |
| Angiogenesis | VEGF, VEGFR1, VEGFR2, COX2 |
| Other pro-oncogenic factors | TNFα, BRCA1, ARHGEF2, Ajuba, MALAT-1, RAS-6TP, FAS |
The pro-oncogenic role of NR4A1 has been investigated in multiple solid tumor derived cancer cell lines (rev. in 70), however, the number of publications on the functions of NR4A1 in pancreatic cancer is limited[55,71,72]. Knockdown of NR4A1 in pancreatic cancer cells inhibits cancer cell and tumor growth in athymic nude mouse xenograft studies, induces apoptosis and inhibits migration/invasion. Thus, the pro-oncogenic functions of NR4A1 and Sp TFs in pancreatic cancer cell lines are similar (Figure 1) and comparable results were obtained after their knockdown. For example, knockdown of NR4A1 by RNA interference in pancreatic cancer cells decreased expression of bcl-2 and survivin and this was accompanied by markers of apoptosis including increased TUNEL staining, caspase-3 and PARP cleavage[55,71]. Proteome analysis confirmed that loss of NR4A1 enhanced cell death and decreased gene products associated with cell proliferation and this was accompanied by increased reactive oxygen species (ROS) and ROS-induced endoplasmic reticulum (ER) stress genes including CHOP, Grp78, ATF4 and XBP-1s[71]. In addition, genes such as thioredoxin domain containing 5 (TXNDC5) and isocitrate dehydrogenase 1 (IDH1) that maintain high reductant levels are also NR4A1 regulated genes and their loss after receptor knockdown triggers the ROS-ER stress response. Knockdown of NR4A1 also decreased cell migration and expression of β1- and α5-integrins which play a role in NR4A1-regulated pancreatic cancer cell migration[72]. Thus, there is overlap between the function of Sp TFs and NR4A1 in pancreatic cancer cell lines and studies in other cancer cell lines show that many of the Sp-regulated genes summarized in Table 1 are also regulated by NR4A1[70]. Regulation of the same genes by Sp TFs and NR4A1 was recently reported in breast cancer cells where knockdown of Sp4 or NR4A1 decreased expression of α6- and β1-integrins in MDA-MB-231 and SKBR3 breast cancer cells. The effects of Sp3 and Sp1 knockdown on expression of these integrins was gene- and cell context-specific.
Both NR4A1 and Sp TFs are nuclear proteins that interact with defined cis-elements in their target gene promoters. Mechanistic studies showing cooperative activation of common genes by NR4A1 and Sp1 was first observed in pancreatic cancer cells and subsequently investigated and confirmed in other cancer cell lines[55,72]. The survivin gene has a GC-rich promoter that binds Sp TFs and has been extensively characterized as an Sp-regulated gene[73,74]. Knockdown of NR4A1 or treatment with NR4A1 antagonists decreased expression of survivin protein and mRNA and also decreased luciferase activity in pancreatic cancer cells transfected with GC-rich survivin promoter-luciferase constructs[55]. Further analysis of survivin regulation in pancreatic cancer cells showed that survivin is regulated by a p300/NR4A1/Sp1 complex where Sp1 binds the GC-rich survivin promoter, NR4A1 acts as a cofactor/coactivator and p300 is also part of the complex. The identification of this functional NR4A1/Sp complex is consistent with previous reports showing many other nuclear receptors also activate gene expression through interactions with Sp1. Studies in other cancer cell lines have identified other NR4A1-regulated genes that are regulated by NR4A1/Sp1, NR4A1/Sp4 or NR4A1/Sp1/4 and these include β1-, β3- and β4-integrins, α5- and α6-integrins, TXNDC5 and the PAX3-FOX01 fusion oncogene and the checkpoint gene PD-L1 (rev. in 70). Thus, like Ajuba[75], NR4A1 acts as a cofactor for Sp TFs in pancreatic and other cancer cell lines and they coregulate some of the same genes (EGFR, survivin and IGF1R). The possible interactions of NR4A1 and Ajuba, a coregulator of Sp-regulated genes needs to be further investigated.
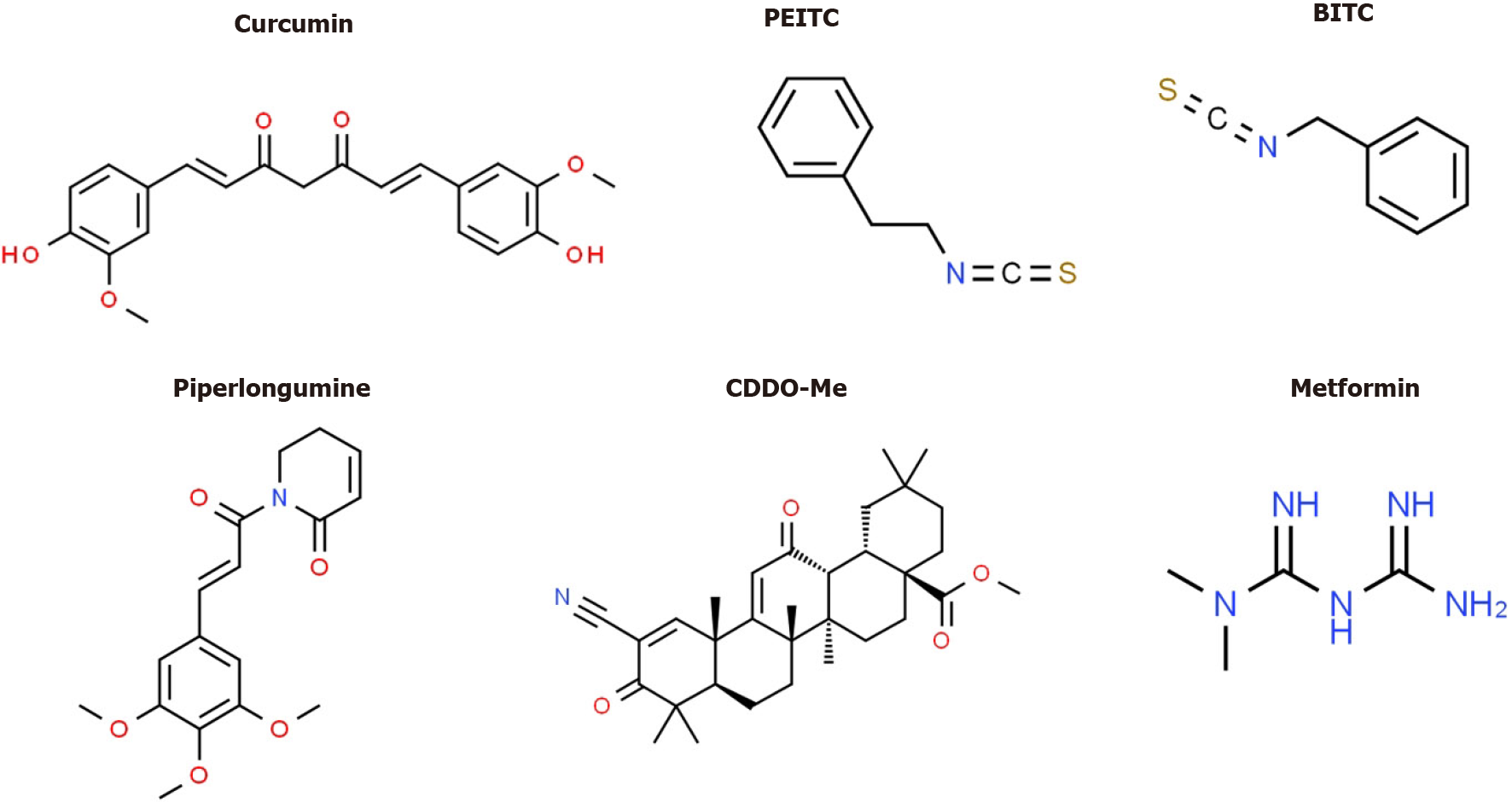
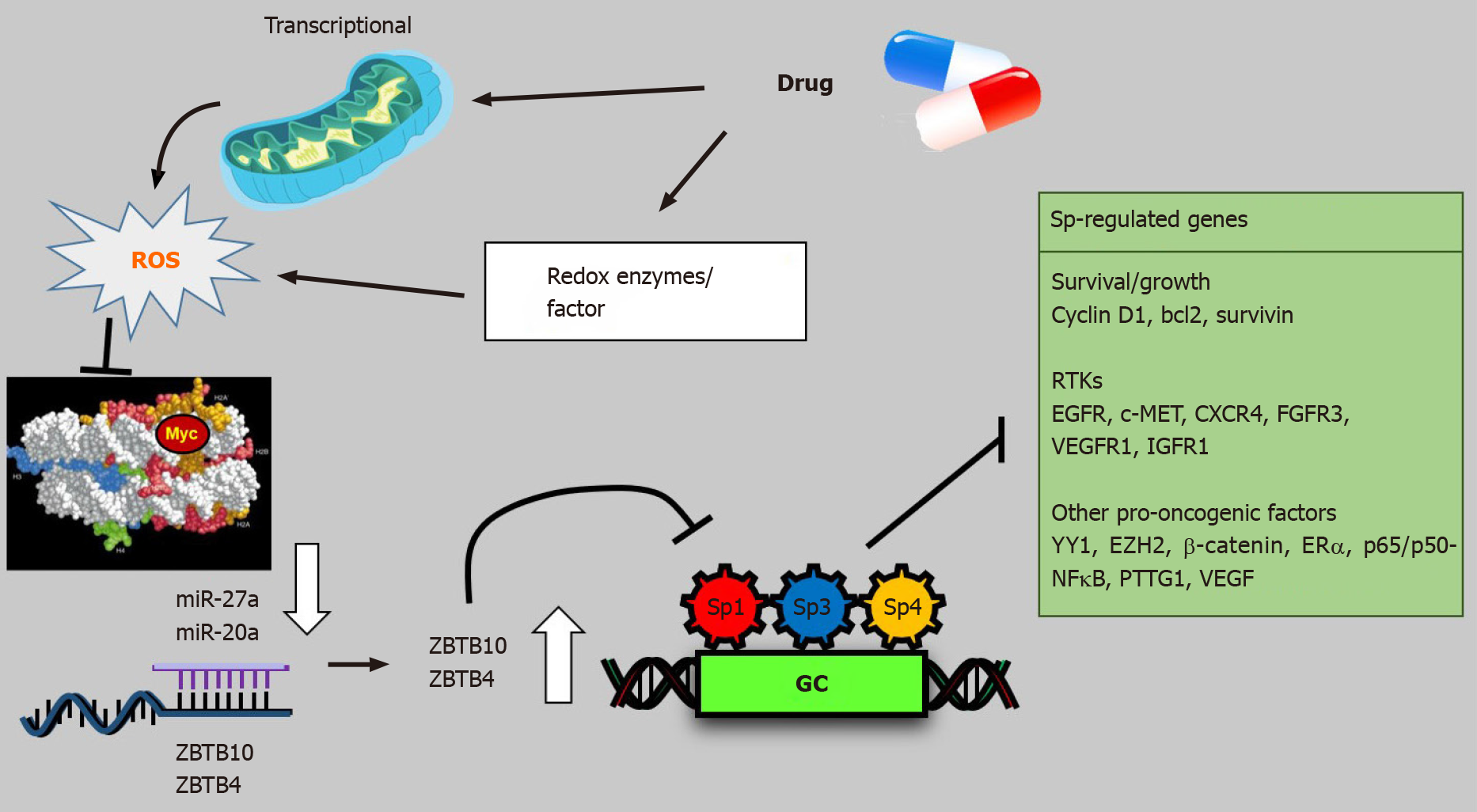
Several drugs that target Sp TFs in pancreatic cancer cells have been identified and these include tolfenamic acid and structurally related compounds, COX2 inhibitors curcumin, metformin, piperlongumine, methyl 2-cyano-3,12-dioxooleana-1,9-dien-28-oate (CDDO-Me), benzylisothiocyanate (BITC) and phenethylisothiocyanate (PEITC) (Figure 3)[20]. All of these compounds downregulate Sp1, Sp3 and Sp4 through different pathways. For example, tolfenamic acid and COX2 inhibitors induce proteasome-dependent degradation of Sp1, Sp3 and Sp4 whereas metformin induces the dual specificity phosphatases MPK-1 and MPK-5 which are important for Sp downregulation. The phosphatases downregulate miR-27a which results in upregulation of ZBTB10, a Sp repressor which competitively displaces Sp TFs from GC-rich sites resulting in gene inactivation since ZBTB10 Lacks a transactivation domain. ROS-inducers including PEITC, BITC, piperlongumine, CDDO-Me and curcumin also decrease Sp-regulated gene expression through an ROS-mediated pathway which also involves induction of Sp repressors namely ZBTB4, ZBTB10 and ZBTB34[20] (Figure 4). This pathway involves ROS-dependent remodeling of repressor complexes which results in downregulation of cMyc and cMyc-dependent microRNAs (miR-27a, miR-17-92) and induction of ZBTBs. These compounds (Figure 4) and others induce similar patterns and mechanisms of Sp downregulation in other cancer cell lines[20], however, there are compound-specific differences in mechanisms that are cell context-dependent.
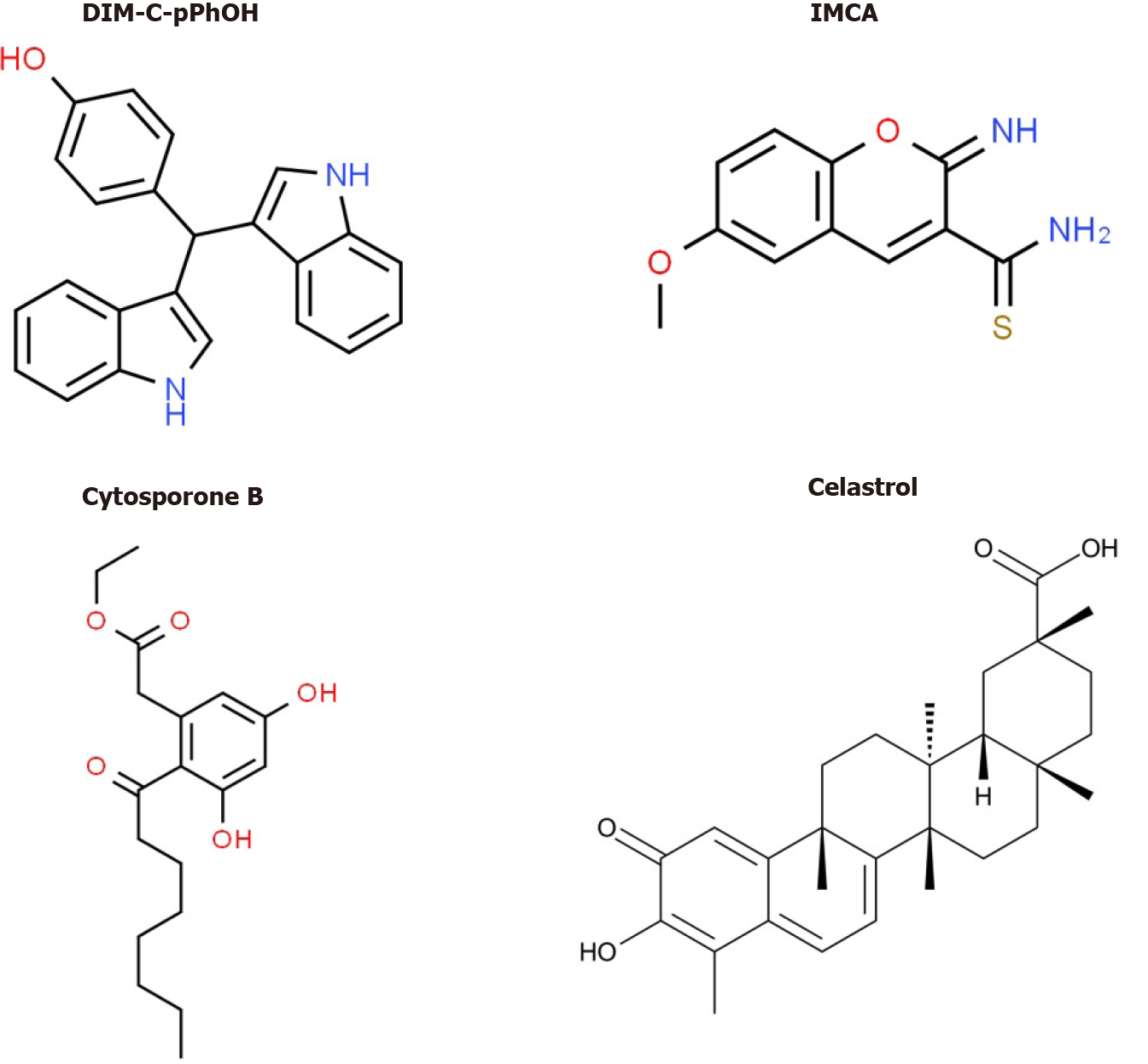
Bis (3΄-indolyl)-1-(p-hydroxyphenyl) methane (DIM-C-pPhOH) (Figure 5) was identified as an NR4A1 Ligand that acts as a receptor antagonist and mimics the effects of NR4A1 knockdown in pancreatic and other cancer cell lines[70]. DIM-C-pPhOH and several 3,5-disubstituted phenyl analogs are inhibitors of cell proliferation, survival and migration/invasion and they inhibit expression of NR4A1/Sp- and other NR4A1-regulated genes. Moreover, many of the 3,5-disubstituted analogs of DIM-C-pPhOH inhibit tumor growth in mouse xenograft models at doses < 5 mg/kg per day and ongoing studies have identified ligands that inhibit tumor growth at doses < 1mg/kg per day. Interestingly, several other classes of compounds bind and modulate NR4A1-dependent pathways/genes and these include cytosporone B and structurally related compounds, IMCA (2-imino-6-methoxy-2H-chromene-3-carbothiomide) and the triterpenoid celastrol and analogs[76-80] (Figure 5). Fascinatingly, celastrol also activates ROS-dependent downregulation of Sp1, Sp3 and Sp4 in bladder cancer cells[80] (e.g., Figure 4) and uniquely acts as a bifunctional agent targeting both Sp TFs and NR4A1. Ongoing studies in this laboratory have identified other dual targeting agents with potential for clinical applications for pancreatic cancer therapy.
In summary, it is evident that both Sp TFs and NR4A1 are pro-oncogenic factors that regulate pancreatic cancer cell and tumor growth and interactions of NR4A1 and Sp TFs are also important for regulating many critical genes (Figure 1) involved in pro-oncogenic functions. Although the functional importance of NR4A1/Sp regulated genes in cancer cell growth, survival, migration and invasion has been established in pancreatic and other cancers, clinical applications of drugs targeting Sp and NR4A1 are lacking. Several compounds have been identified that induce Sp downregulation or inhibit/antagonize NR4A1. Current studies in our laboratories are focused on identifying agents like celastrol that act simultaneously as NR4A1 antagonists and Sp downregulators as a new class of drugs that will enhance the effectiveness of current chemotherapies used for clinical treatment of pancreatic and other cancers.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Li JT, Matsuo Y, Uhlmann D S-Editor: Gao CC L-Editor: A P-Editor: Liu JH
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12667] [Cited by in F6Publishing: 13952] [Article Influence: 3488.0] [Reference Citation Analysis (4)] |
| 2. | Zhang Y, Yang C, Cheng H, Fan Z, Huang Q, Lu Y, Fan K, Luo G, Jin K, Wang Z, Liu C, Yu X. Novel agents for pancreatic ductal adenocarcinoma: emerging therapeutics and future directions. J Hematol Oncol. 2018;11:14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 3. | Karandish F, Mallik S. Biomarkers and Targeted Therapy in Pancreatic Cancer. Biomark Cancer. 2016;8:27-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 4. | Michael JV, Goldfinger LE. Concepts and advances in cancer therapeutic vulnerabilities in RAS membrane targeting. Semin Cancer Biol. 2019;54:121-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111:817-822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 365] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 6. | Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801-1806. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3082] [Cited by in F6Publishing: 2897] [Article Influence: 181.1] [Reference Citation Analysis (0)] |
| 7. | Qian Y, Gong Y, Fan Z, Luo G, Huang Q, Deng S, Cheng H, Jin K, Ni Q, Yu X, Liu C. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J Hematol Oncol. 2020;13:130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 152] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 8. | Stoica AF, Chang CH, Pauklin S. Molecular Therapeutics of Pancreatic Ductal Adenocarcinoma: Targeted Pathways and the Role of Cancer Stem Cells. Trends Pharmacol Sci. 2020;41:977-993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Isacoff WH, Reber HA, Bedford R, Hoos W, Rahib L, Upfill-Brown A, Donahue T, Hines OJ. Low-Dose Continuous 5-Fluorouracil Combined with Leucovorin, nab-Paclitaxel, Oxaliplatin, and Bevacizumab for Patients with Advanced Pancreatic Cancer: A Retrospective Analysis. Target Oncol. 2018;13:461-468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 294] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 11. | Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619-628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 386] [Cited by in F6Publishing: 412] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 12. | Vizcaíno C, Mansilla S, Portugal J. Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol Ther. 2015;152:111-124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 250] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 13. | Beishline K, Azizkhan-Clifford J. Sp1 and the 'hallmarks of cancer'. FEBS J. 2015;282:224-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 368] [Cited by in F6Publishing: 353] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 14. | Kim CK, He P, Bialkowska AB, Yang VW. SP and KLF Transcription Factors in Digestive Physiology and Diseases. Gastroenterology. 2017;152:1845-1875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 15. | Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079-1090. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1188] [Cited by in F6Publishing: 1350] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 16. | Oh JE, Han JA, Hwang ES. Downregulation of transcription factor, Sp1, during cellular senescence. Biochem Biophys Res Commun. 2007;353:86-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Adrian GS, Seto E, Fischbach KS, Rivera EV, Adrian EK, Herbert DC, Walter CA, Weaker FJ, Bowman BH. YY1 and Sp1 transcription factors bind the human transferrin gene in an age-related manner. J Gerontol A Biol Sci Med Sci. 1996;51:B66-B75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Park SC. Nuclear barrier hypothesis of aging as mechanism for trade-off growth to survival. Adv Exp Med Biol. 2011;720:3-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 19. | Kim SY, Kang HT, Han JA, Park SC. The transcription factor Sp1 is responsible for aging-dependent altered nucleocytoplasmic trafficking. Aging Cell. 2012;11:1102-1109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Safe S, Abbruzzese J, Abdelrahim M, Hedrick E. Specificity Protein Transcription Factors and Cancer: Opportunities for Drug Development. Cancer Prev Res (Phila). 2018;11:371-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 21. | Jiang NY, Woda BA, Banner BF, Whalen GF, Dresser KA, Lu D. Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:1648-1652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 120] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 22. | Kong LM, Liao CG, Fei F, Guo X, Xing JL, Chen ZN. Transcription factor Sp1 regulates expression of cancer-associated molecule CD147 in human lung cancer. Cancer Sci. 2010;101:1463-1470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 23. | Li L, Gao P, Li Y, Shen Y, Xie J, Sun D, Xue A, Zhao Z, Xu Z, Zhang M, Li B, Jiang J. JMJD2A-dependent silencing of Sp1 in advanced breast cancer promotes metastasis by downregulation of DIRAS3. Breast Cancer Res Treat. 2014;147:487-500. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 24. | Wang XB, Peng WQ, Yi ZJ, Zhu SL, Gan QH. [Expression and prognostic value of transcriptional factor sp1 in breast cancer]. Ai Zheng. 2007;26:996-1000. [PubMed] [Cited in This Article: ] |
| 25. | Kim JY, Jung HH, Ahn S, Bae S, Lee SK, Kim SW, Lee JE, Nam SJ, Ahn JS, Im YH, Park YH. The relationship between nuclear factor (NF)-κB family gene expression and prognosis in triple-negative breast cancer (TNBC) patients receiving adjuvant doxorubicin treatment. Sci Rep. 2016;6:31804. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Wang L, Wei D, Huang S, Peng Z, Le X, Wu TT, Yao J, Ajani J, Xie K. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371-6380. [PubMed] [Cited in This Article: ] |
| 27. | Lee HS, Park CK, Oh E, Erkin ÖC, Jung HS, Cho MH, Kwon MJ, Chae SW, Kim SH, Wang LH, Park MJ, Lee SY, Yang HB, Jia L, Choi YL, Shin YK. Low SP1 expression differentially affects intestinal-type compared with diffuse-type gastric adenocarcinoma. PLoS One. 2013;8:e55522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Yao JC, Wang L, Wei D, Gong W, Hassan M, Wu TT, Mansfield P, Ajani J, Xie K. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin Cancer Res. 2004;10:4109-4117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 159] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 29. | Guan H, Cai J, Zhang N, Wu J, Yuan J, Li J, Li M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int J Cancer. 2012;130:593-601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 30. | Dong Q, Cai N, Tao T, Zhang R, Yan W, Li R, Zhang J, Luo H, Shi Y, Luan W, Zhang Y, You Y, Wang Y, Liu N. An axis involving SNAI1, microRNA-128 and SP1 modulates glioma progression. PLoS One. 2014;9:e98651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 31. | Maurer GD, Leupold JH, Schewe DM, Biller T, Kates RE, Hornung HM, Lau-Werner U, Post S, Allgayer H. Analysis of specific transcriptional regulators as early predictors of independent prognostic relevance in resected colorectal cancer. Clin Cancer Res. 2007;13:1123-1132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 32. | Chen YT, Tsai HP, Wu CC, Chen CY, Chai CY, Kwan AL. High-level Sp1 is Associated with Proliferation, Invasion, and Poor Prognosis in Astrocytoma. Pathol Oncol Res. 2019;25:1003-1013. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Liu L, Ji P, Qu N, Pu WL, Jiang DW, Liu WY, Li YQ, Shi RL. The impact of high co-expression of Sp1 and HIF1α on prognosis of patients with hepatocellular cancer. Oncol Lett. 2016;12:504-512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Kong LM, Yao L, Lu N, Dong YL, Zhang J, Wang YQ, Liu L, Zhang HL, Huang JG, Liao CG. Interaction of KLF6 and Sp1 regulates basigin-2 expression mediated proliferation, invasion and metastasis in hepatocellular carcinoma. Oncotarget. 2016;7:27975-27987. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Hu J, Hu H, Hang JJ, Yang HY, Wang ZY, Wang L, Chen DH, Wang LW. Simultaneous high expression of PLD1 and Sp1 predicts a poor prognosis for pancreatic ductal adenocarcinoma patients. Oncotarget. 2016;7:78557-78565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Zhang HW, Wang EW, Li LX, Yi SH, Li LC, Xu FL, Wang DL, Wu YZ, Nian WQ. A regulatory loop involving miR-29c and Sp1 elevates the TGF-β1 mediated epithelial-to-mesenchymal transition in lung cancer. Oncotarget. 2016;7:85905-85916. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 37. | Zhang J, Zhu ZG, Ji J, Yuan F, Yu YY, Liu BY, Lin YZ. Transcription factor Sp1 expression in gastric cancer and its relationship to long-term prognosis. World J Gastroenterol. 2005;11:2213-2217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 35] [Cited by in F6Publishing: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 38. | Essafi-Benkhadir K, Grosso S, Puissant A, Robert G, Essafi M, Deckert M, Chamorey E, Dassonville O, Milano G, Auberger P, Pagès G. Dual role of Sp3 transcription factor as an inducer of apoptosis and a marker of tumour aggressiveness. PLoS One. 2009;4:e4478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Bedolla RG, Gong J, Prihoda TJ, Yeh IT, Thompson IM, Ghosh R, Kumar AP. Predictive value of Sp1/Sp3/FLIP signature for prostate cancer recurrence. PLoS One. 2012;7:e44917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC, Chang WC, Hung JJ. Sp1 expression regulates lung tumor progression. Oncogene. 2012;31:3973-3988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 41. | Lou Z, O'Reilly S, Liang H, Maher VM, Sleight SD, McCormick JJ. Down-regulation of overexpressed sp1 protein in human fibrosarcoma cell lines inhibits tumor formation. Cancer Res. 2005;65:1007-1017. [PubMed] [Cited in This Article: ] |
| 42. | Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 297] [Cited by in F6Publishing: 322] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 43. | Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891-1903. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 217] [Cited by in F6Publishing: 236] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 44. | Lee SL, Wesselschmidt RL, Linette GP, Kanagawa O, Russell JH, Milbrandt J. Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77). Science. 1995;269:532-535. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 222] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 45. | Zetterström RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 842] [Cited by in F6Publishing: 846] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 46. | Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A. 1998;95:4013-4018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 586] [Cited by in F6Publishing: 581] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 47. | Cheng LE, Chan FK, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 1997;16:1865-1875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 247] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 48. | Woronicz JD, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994;367:277-281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 449] [Cited by in F6Publishing: 465] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 49. | Wilson TE, Fahrner TJ, Johnston M, Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991;252:1296-1300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 421] [Cited by in F6Publishing: 446] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 50. | Maira M, Martens C, Philips A, Drouin J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol. 1999;19:7549-7557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 189] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 51. | Philips A, Lesage S, Gingras R, Maira MH, Gauthier Y, Hugo P, Drouin J. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol Cell Biol. 1997;17:5946-5951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 267] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 52. | Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 1995;9:769-782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 405] [Cited by in F6Publishing: 415] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 53. | Zetterström RH, Solomin L, Mitsiadis T, Olson L, Perlmann T. Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol Endocrinol. 1996;10:1656-1666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 57] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 54. | Kurakula K, Koenis DS, van Tiel CM, de Vries CJ. NR4A nuclear receptors are orphans but not lonesome. Biochim Biophys Acta. 2014;1843:2543-2555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 103] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 55. | Lee SO, Abdelrahim M, Yoon K, Chintharlapalli S, Papineni S, Kim K, Wang H, Safe S. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Res. 2010;70:6824-6836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 56. | Lacey A, Rodrigues-Hoffman A, Safe S. PAX3-FOXO1A Expression in Rhabdomyosarcoma Is Driven by the Targetable Nuclear Receptor NR4A1. Cancer Res. 2017;77:732-741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 57. | Smith AG, Lim W, Pearen M, Muscat GE, Sturm RA. Regulation of NR4A nuclear receptor expression by oncogenic BRAF in melanoma cells. Pigment Cell Melanoma Res. 2011;24:551-563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 58. | Wang JR, Gan WJ, Li XM, Zhao YY, Li Y, Lu XX, Li JM, Wu H. Orphan nuclear receptor Nur77 promotes colorectal cancer invasion and metastasis by regulating MMP-9 and E-cadherin. Carcinogenesis. 2014;35:2474-2484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 59. | Zhou F, Drabsch Y, Dekker TJ, de Vinuesa AG, Li Y, Hawinkels LJ, Sheppard KA, Goumans MJ, Luwor RB, de Vries CJ, Mesker WE, Tollenaar RA, Devilee P, Lu CX, Zhu H, Zhang L, Dijke PT. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nat Commun. 2014;5:3388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 60. | Delgado E, Boisen MM, Laskey R, Chen R, Song C, Sallit J, Yochum ZA, Andersen CL, Sikora MJ, Wagner J, Safe S, Elishaev E, Lee A, Edwards RP, Haluska P, Tseng G, Schurdak M, Oesterreich S. High expression of orphan nuclear receptor NR4A1 in a subset of ovarian tumors with worse outcome. Gynecol Oncol. 2016;141:348-356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 61. | Zhu B, Yang JR, Jia Y, Zhang P, Shen L, Li XL, Li J, Wang B. Overexpression of NR4A1 is associated with tumor recurrence and poor survival in non-small-cell lung carcinoma. Oncotarget. 2017;8:113977-113986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 62. | Muscat GE, Eriksson NA, Byth K, Loi S, Graham D, Jindal S, Davis MJ, Clyne C, Funder JW, Simpson ER, Ragan MA, Kuczek E, Fuller PJ, Tilley WD, Leedman PJ, Clarke CL. Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Mol Endocrinol. 2013;27:350-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 63. | Hu Y, Chau T, Liu HX, Liao D, Keane R, Nie Y, Yang H, Wan YJ. Bile acids regulate nuclear receptor (Nur77) expression and intracellular location to control proliferation and apoptosis. Mol Cancer Res. 2015;13:281-292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 64. | Cho HJ, Zhao J, Jung SW, Ladewig E, Kong DS, Suh YL, Lee Y, Kim D, Ahn SH, Bordyuh M, Kang HJ, Sa JK, Seo YJ, Kim ST, Lim DH, Dho YS, Lee JI, Seol HJ, Choi JW, Park WY, Park CK, Rabadan R, Nam DH. Distinct genomic profile and specific targeted drug responses in adult cerebellar glioblastoma. Neuro Oncol. 2019;21:47-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 65. | Shi Q, Le X, Abbruzzese JL, Peng Z, Qian CN, Tang H, Xiong Q, Wang B, Li XC, Xie K. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 2001;61:4143-4154. [PubMed] [Cited in This Article: ] |
| 66. | Hedrick E, Cheng Y, Jin UH, Kim K, Safe S. Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 are non-oncogene addiction genes in cancer cells. Oncotarget. 2016;7:22245-22256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 67. | Kent OA, Mendell JT, Rottapel R. Transcriptional Regulation of miR-31 by Oncogenic KRAS Mediates Metastatic Phenotypes by Repressing RASA1. Mol Cancer Res. 2016;14:267-277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 68. | Li S, Wang Q, Qiang Q, Shan H, Shi M, Chen B, Zhao S, Yuan L. Sp1-mediated transcriptional regulation of MALAT1 plays a critical role in tumor. J Cancer Res Clin Oncol. 2015;141:1909-1920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 69. | Tan Y, Yin H, Zhang H, Fang J, Zheng W, Li D, Li Y, Cao W, Sun C, Liang Y, Zeng J, Zou H, Fu W, Yang X. Sp1-driven up-regulation of miR-19a decreases RHOB and promotes pancreatic cancer. Oncotarget. 2015;6:17391-17403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 70. | Safe S, Karki K. The Paradoxical Roles of Orphan Nuclear Receptor 4A (NR4A) in Cancer. Mol Cancer Res. 2021;19:180-191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 71. | Lee SO, Jin UH, Kang JH, Kim SB, Guthrie AS, Sreevalsan S, Lee JS, Safe S. The orphan nuclear receptor NR4A1 (Nur77) regulates oxidative and endoplasmic reticulum stress in pancreatic cancer cells. Mol Cancer Res. 2014;12:527-538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 72. | Hedrick E, Lee SO, Safe S. The nuclear orphan receptor NR4A1 regulates β1-integrin expression in pancreatic and colon cancer cells and can be targeted by NR4A1 antagonists. Mol Carcinog. 2017;56:2066-2075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 73. | Stauber RH, Mann W, Knauer SK. Nuclear and cytoplasmic survivin: molecular mechanism, prognostic, and therapeutic potential. Cancer Res. 2007;67:5999-6002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 175] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 74. | Wu J, Ling X, Pan D, Apontes P, Song L, Liang P, Altieri DC, Beerman T, Li F. Molecular mechanism of inhibition of survivin transcription by the GC-rich sequence-selective DNA binding antitumor agent, hedamycin: evidence of survivin down-regulation associated with drug sensitivity. J Biol Chem. 2005;280:9745-9751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 75. | Zhang B, Song L, Cai J, Li L, Xu H, Li M, Wang J, Shi M, Chen H, Jia H, Hou Z. The LIM protein Ajuba/SP1 complex forms a feed forward loop to induce SP1 target genes and promote pancreatic cancer cell proliferation. J Exp Clin Cancer Res. 2019;38:205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 76. | Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, Li G, Xu Q, Zhang M, Weimer BC, Chen D, Cheng Z, Zhang L, Li Q, Li S, Zheng Z, Song S, Huang Y, Ye Z, Su W, Lin SC, Shen Y, Wu Q. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol. 2008;4:548-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 248] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 77. | Wang WJ, Wang Y, Chen HZ, Xing YZ, Li FW, Zhang Q, Zhou B, Zhang HK, Zhang J, Bian XL, Li L, Liu Y, Zhao BX, Chen Y, Wu R, Li AZ, Yao LM, Chen P, Zhang Y, Tian XY, Beermann F, Wu M, Han J, Huang PQ, Lin T, Wu Q. Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway. Nat Chem Biol. 2014;10:133-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 150] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 78. | Zhang L, Liu W, Wang Q, Li Q, Wang H, Wang J, Teng T, Chen M, Ji A, Li Y. New Drug Candidate Targeting the 4A1 Orphan Nuclear Receptor for Medullary Thyroid Cancer Therapy. Molecules. 2018;23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 79. | Hu M, Luo Q, Alitongbieke G, Chong S, Xu C, Xie L, Chen X, Zhang D, Zhou Y, Wang Z, Ye X, Cai L, Zhang F, Chen H, Jiang F, Fang H, Yang S, Liu J, Diaz-Meco MT, Su Y, Zhou H, Moscat J, Lin X, Zhang XK. Celastrol-Induced Nur77 Interaction with TRAF2 Alleviates Inflammation by Promoting Mitochondrial Ubiquitination and Autophagy. Mol Cell. 2017;66:141-153.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 191] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 80. | Chen Z, Zhang D, Yan S, Hu C, Huang Z, Li Z, Peng S, Li X, Zhu Y, Yu H, Lian B, Kang Q, Li M, Zeng Z, Zhang XK, Su Y. SAR study of celastrol analogs targeting Nur77-mediated inflammatory pathway. Eur J Med Chem. 2019;177:171-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |