Copyright
©The Author(s) 2021.
World J Gastroenterol. Jun 21, 2021; 27(23): 3182-3207
Published online Jun 21, 2021. doi: 10.3748/wjg.v27.i23.3182
Published online Jun 21, 2021. doi: 10.3748/wjg.v27.i23.3182
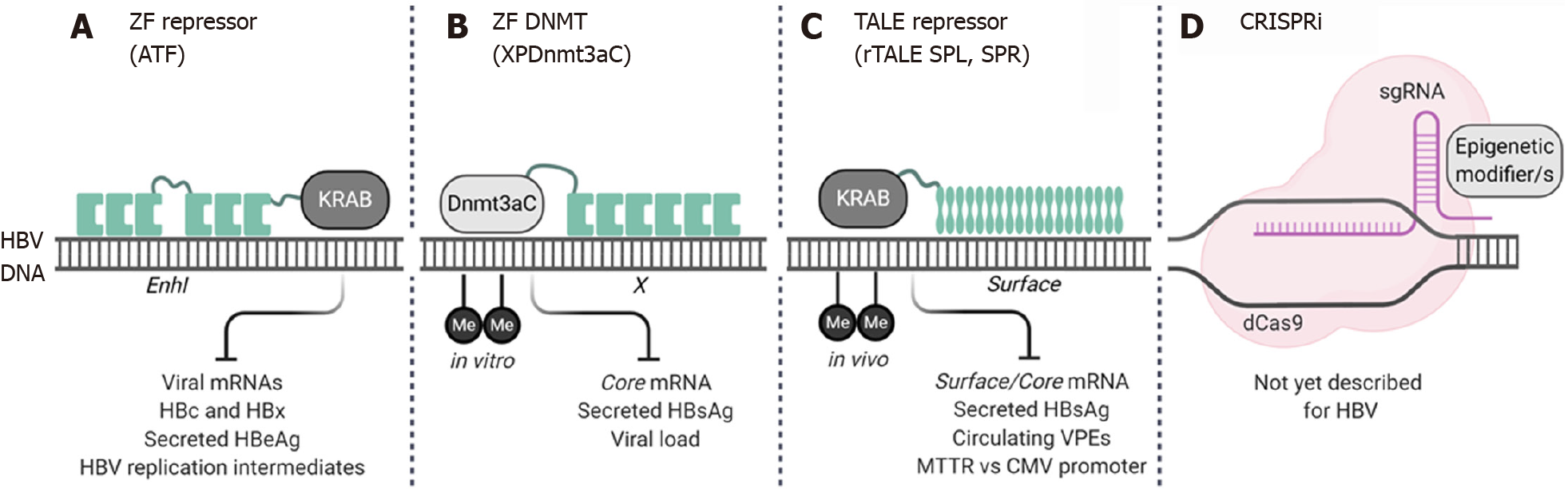
Figure 2 Novel epigenome engineering tools that target hepatitis B virus DNA.
Three different types of epigenetic editors have been designed to silence viral gene expression. The Zinc finger (ZF) repressor-based artificial transcription factors (ATF) comprise two three-finger ZF DNA binding domains (DBD) which are fused, with a C-terminal linker peptide, to the Krüppelassociated box (KRAB) repressor domain. ATFs were designed to target the EnhI region (A). A ZF-based methyltransferase (DNMT) designed to target the X region (XPDnmt3aC). The XPDnmt3aC comprises a six-finger ZF DBD fused to the catalytic domain of Dnmt3a (Dnmt3aC). Methylation of the viral DNA was achieved in vitro (B). To generate transcription activator-like effector (TALE) repressors, TALE DBDs targeting two regions of surface were fused to the KRAB repressor domain (N-terminal). In vivo, methylation of intrahepatic viral DNA was shown (C). Although not yet described for HBV, the CRISPRi epigenome editing tools provide a novel means of silencing covalently closed circular DNA. RNA guides (sgRNA) targeting the viral genome could be used to recruit a dead Cas (dCas9) with one or more epigenetic modifying domains (D). Created with BioRender.com. HBV: Hepatitis B virus; ZF: Zinc finger; ATF: Artificial transcription factor; HBx: Hepatitis B virus X protein; HBc: Hepatitis B virus core protein; HBeAg: Hepatitis B e antigen; HBsAg: Hepatitis B surface antigen; TALE: Transcription activator-like effector; MTTR: Mouse transthyretin; CMV: Cytomegalovirus; VPE: Viral particle equivalent.
- Citation: Singh P, Kairuz D, Arbuthnot P, Bloom K. Silencing hepatitis B virus covalently closed circular DNA: The potential of an epigenetic therapy approach. World J Gastroenterol 2021; 27(23): 3182-3207
- URL: https://www.wjgnet.com/1007-9327/full/v27/i23/3182.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i23.3182