Copyright
©The Author(s) 2020.
World J Gastroenterol. Oct 21, 2020; 26(39): 5983-5996
Published online Oct 21, 2020. doi: 10.3748/wjg.v26.i39.5983
Published online Oct 21, 2020. doi: 10.3748/wjg.v26.i39.5983
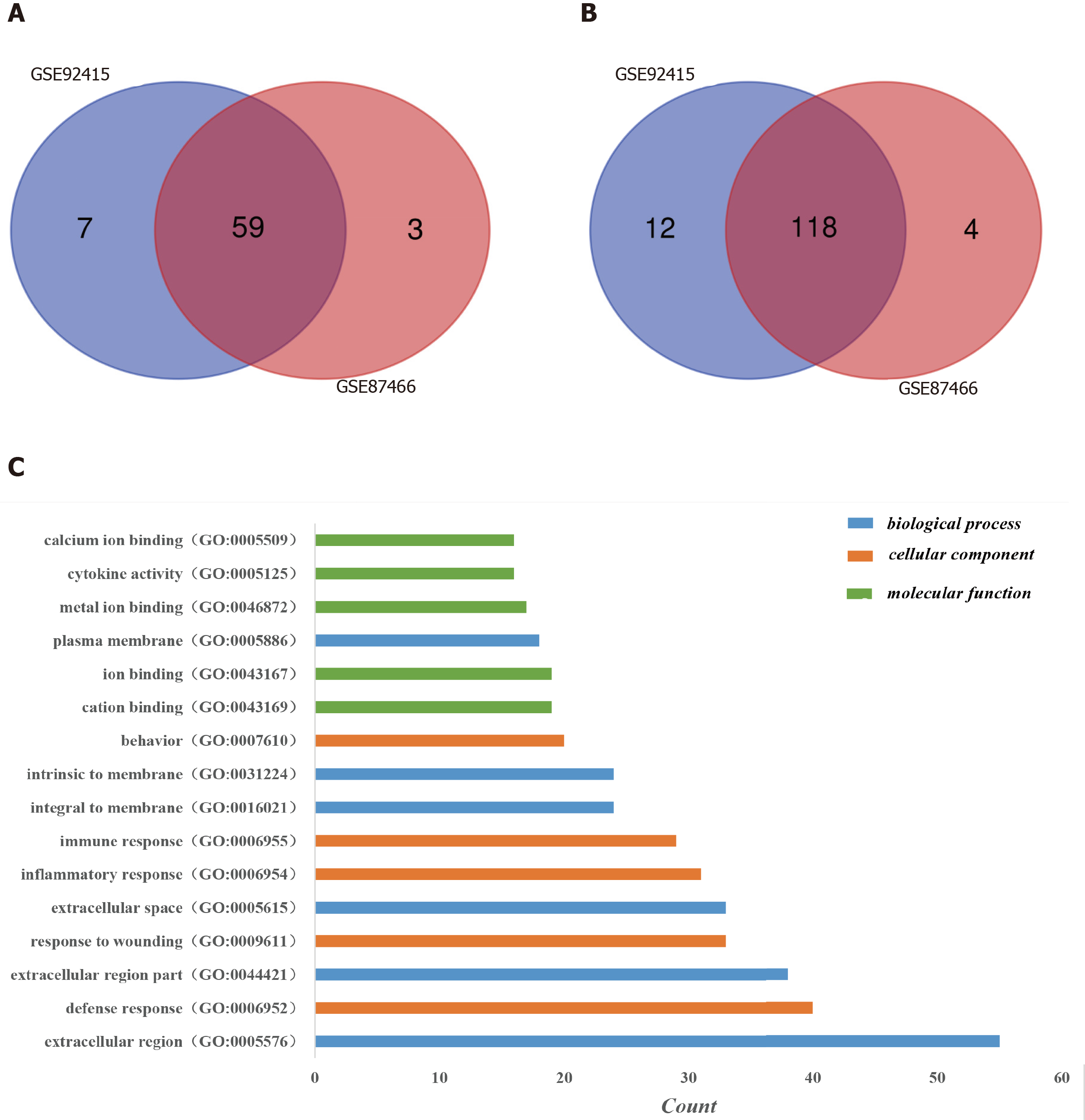
Figure 1 Identification of differentially expressed genes in the two databases (GSE92415 and GSE87466) and Gene Ontology analysis of differentially expressed genes in ulcerative colitis.
A: Upregulated differentially expressed genes (DEGs); B: Downregulated DEGs. DEGs were identified with a t-test, and statistically significant DEGs were defined by the GEO2R online tool with a |logFC| > 2 and adjusted P value < 0.05; C: The Gene Ontology analysis classified the DEGs into three groups: Molecular function, biological process and cellular component. Terms were selected with > 15 genes and arrayed in ascending order from top to bottom according to the count. GO: Gene Ontology.
- Citation: Shi L, Han X, Li JX, Liao YT, Kou FS, Wang ZB, Shi R, Zhao XJ, Sun ZM, Hao Y. Identification of differentially expressed genes in ulcerative colitis and verification in a colitis mouse model by bioinformatics analyses. World J Gastroenterol 2020; 26(39): 5983-5996
- URL: https://www.wjgnet.com/1007-9327/full/v26/i39/5983.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i39.5983