Copyright
©The Author(s) 2019.
World J Gastroenterol. Aug 28, 2019; 25(32): 4661-4672
Published online Aug 28, 2019. doi: 10.3748/wjg.v25.i32.4661
Published online Aug 28, 2019. doi: 10.3748/wjg.v25.i32.4661
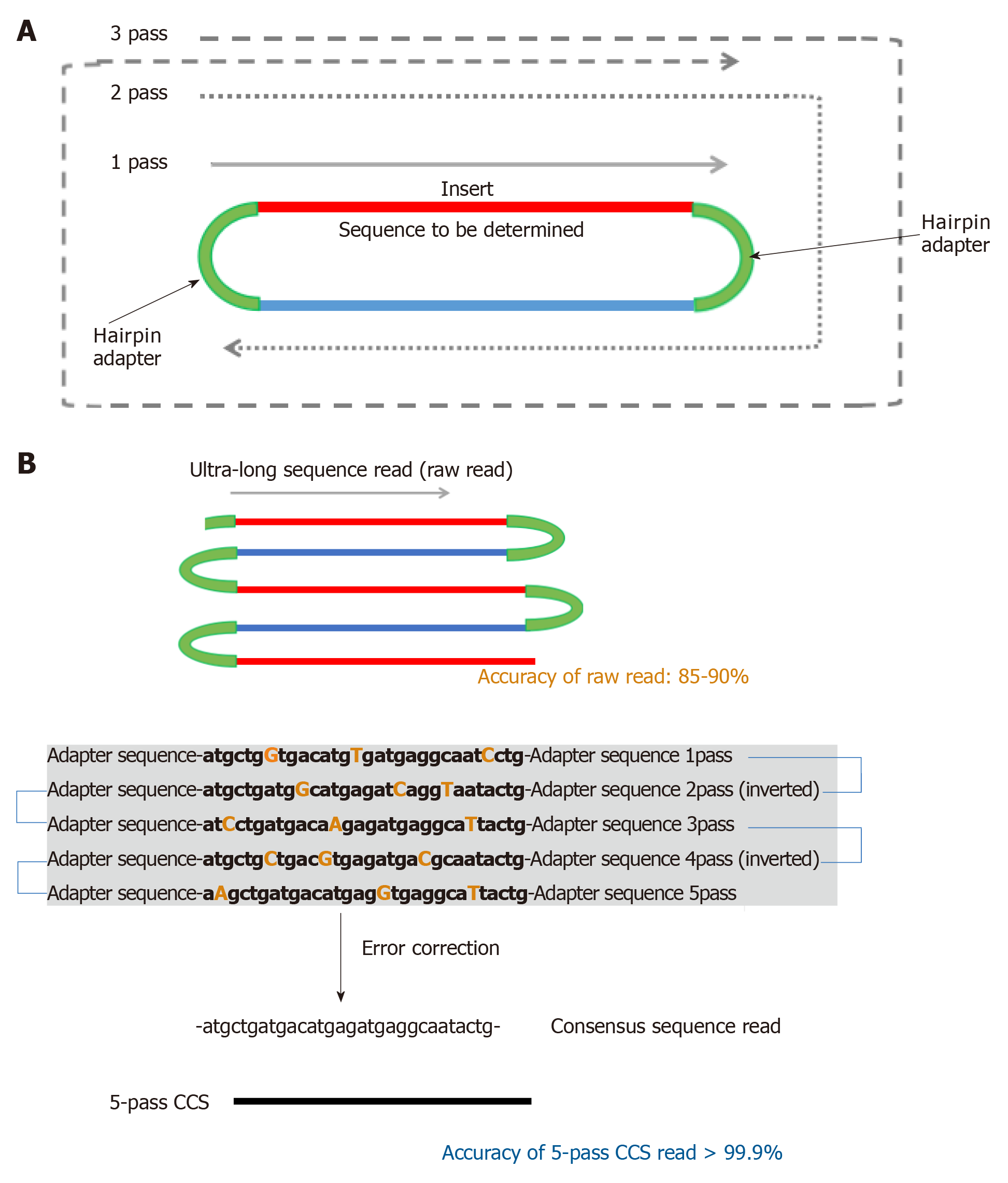
Figure 2 Generation of circular consensus sequences.
A: The template for PacBio sequencing, called SMRTBell, is created by ligating hairpin adaptors to both ends of a double-stranded DNA molecule containing the sequence to be determined. This template then acts like a single-stranded closed circle. The polymerase initiates at the primer location and sequences the template until it falls off. The enzyme then proceeeds around the hairpin on the other end of the SMRTBell, and can circle around the same template multiple times; B: Scheme for generation of 5-pass circular consensus sequences (CCS) reads. Ultra-long raw reads are generated by a polymerase. Although the accuracy of the raw read is 85%-90%, error-corrected consensus reads (CCS reads) can be generated using the data from a single template sequenced multiple times. The accuracy of 5-pass CCS reads is as high as 99.9%. CCS: Circular consensus sequences.
- Citation: Takeda H, Yamashita T, Ueda Y, Sekine A. Exploring the hepatitis C virus genome using single molecule real-time sequencing. World J Gastroenterol 2019; 25(32): 4661-4672
- URL: https://www.wjgnet.com/1007-9327/full/v25/i32/4661.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i32.4661