Published online May 7, 2019. doi: 10.3748/wjg.v25.i17.2086
Peer-review started: January 10, 2019
First decision: February 26, 2019
Revised: March 20, 2019
Accepted: April 10, 2019
Article in press: April 10, 2019
Published online: May 7, 2019
Pancreatic cancer is a highly invasive malignant tumor. Expression levels of the autophagy-related protein microtubule-associated protein 1A/1B-light chain 3 (LC3) and perineural invasion (PNI) are closely related to its occurrence and development. Our previous results showed that the high expression of LC3 was positively correlated with PNI in the patients with pancreatic cancer. In this study, we further searched for differential genes involved in autophagy of pancreatic cancer by gene expression profiling and analyzed their biological functions in pancreatic cancer, which provides a theoretical basis for elucidating the pathophysiological mechanism of autophagy in pancreatic cancer and PNI.
To identify differentially expressed genes involved in pancreatic cancer autophagy and explore the pathogenesis at the molecular level.
Two sets of gene expression profiles of pancreatic cancer/normal tissue (GSE16515 and GSE15471) were collected from the Gene Expression Omnibus. Significance analysis of microarrays algorithm was used to screen differentially expressed genes related to pancreatic cancer. Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were used to analyze the functional enrichment of the differentially expressed genes. Protein interaction data containing only differentially expressed genes was downloaded from String database and screened. Module mining was carried out by Cytoscape software and ClusterOne plug-in. The interaction relationship between the modules was analyzed and the pivot nodes between the functional modules were determined according to the information of the functional modules and the data of reliable protein interaction network.
Based on the above two data sets of pancreatic tissue total gene expression, 6098 and 12928 differentially expressed genes were obtained by analysis of genes with higher phenotypic correlation. After extracting the intersection of the two differential gene sets, 4870 genes were determined. GO analysis showed that 14 significant functional items including negative regulation of protein ubiquitination were closely related to autophagy. A total of 986 differentially expressed genes were enriched in these functional items. After eliminating the autophagy related genes of human cancer cells which had been defined, 347 differentially expressed genes were obtained. KEGG pathway analysis showed that the pathways hsa04144 and hsa04020 were related to autophagy. In addition, 65 clustering modules were screened after the protein interaction network was constructed based on String database, and module 32 contains the LC3 gene, which interacts with multiple autophagy-related genes. Moreover, ubiquitin C acts as a pivot node in functional modules to connect multiple modules related to pancreatic cancer and autophagy.
Three hundred and forty-seven genes associated with autophagy in human pancreatic cancer were concentrated, and a key gene ubiquitin C which is closely related to the occurrence of PNI was determined, suggesting that LC3 may influence the PNI and prognosis of pancreatic cancer through ubiquitin C.
Core tip: In this study, we identified differentially expressed genes based on the autophagy-related protein microtubule-associated protein 1A/1B-light chain 3 (LC3) to analyze the gene expression profile of autophagy in pancreatic cancer. Three hundred and forty-seven genes that have no confirmed association with the autophagy process of human pancreatic cancer cells in previous studies were concentrated, and the key pathways involved in autophagy were enriched. Furthermore, a key gene ubiquitin C which is closely related to the occurrence of perineural invasion (PNI) was determined, suggesting that LC3 may influence the PNI and prognosis of pancreatic cancer through ubiquitin C.
- Citation: Yang YH, Zhang YX, Gui Y, Liu JB, Sun JJ, Fan H. Analysis of the autophagy gene expression profile of pancreatic cancer based on autophagy-related protein microtubule-associated protein 1A/1B-light chain 3. World J Gastroenterol 2019; 25(17): 2086-2098
- URL: https://www.wjgnet.com/1007-9327/full/v25/i17/2086.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i17.2086
Pancreatic cancer is a highly invasive tumor of the digestive system. It is of high malignancy, its early diagnosis is difficult, and it is not sensitive to radiotherapy and chemotherapy. At present, surgical resection is the only relatively effective treatment. The majority of patients are at the late stage of the disease when diagnosed and thus have missed the best treatment opportunity[1]. Therefore, identifying a new direction for the treatment of pancreatic cancer has become the focus of pancreatic surgery studies. As an important mechanism for tumor cells to escape apoptosis, autophagy has both promoting and inhibiting effects on tumors[2,3]. At present, more and more studies have found that autophagy is closely related to the occurrence, development, differentiation and prognosis of pancreatic cancer[4,5].
Autophagy-related protein microtubule-associated protein 1A/1B-light chain 3 (LC3), as a key protein in the autophagy process, is involved in the formation of the autophagosome. A study by our research group found that high expression of LC3 in pancreatic cancer was positively correlated with neural invasion and poor prognosis[6]. On the basis of previous studies and using LC3 as a guidance index, the autophagy gene expression profile of pancreatic cancer was analyzed to guide the functional annotation of differentially expressed genes and to enhance the reliability of bioinformatics prediction and analysis. Differentially expressed genes involved in the autophagy of pancreatic cancer were identified by a gene expression microarray technique. Protein interaction networks were constructed and the functional clustering of differentially expressed genes was carried out. Key interacting proteins or genes between modules were screened and evaluated by statistical methods, and the pathogenesis of pancreatic cancer was explored.
Two groups of experimental data were selected for the analysis of the whole-genome expression profile. Group 1 consisted of 16 normal pancreatic tissue samples and 36 pancreatic cancer tissue samples. Group 2 consisted of 26 normal pancreatic tissue samples and 26 pancreatic cancer tissue samples. All samples were obtained from surgical excision specimens of pancreatic cancer patients and were diagnosed, classified, graded and staged by pathology professionals.
Following the instructions of the Affymetrix gene microarray expression analysis manual and using the Affymetrix Human Genome U133 Plus 2.0 Array sequencing platform (platform number: GPL570)[7-10], gene expression in the samples was examined. All of the obtained data have been uploaded and submitted to the Gene Expression Omnibus (GEO), a gene expression database.
Based on the information of the above two sets of samples and by exploring the gene expression database of the National Center for Biotechnology Information (NCBI) of the United States[11], two sets of pancreatic tissue gene expression profiles were collected: GSE16515 and GSE15471. Data set-related information is listed in Table 1.
| Data set number(raw data of CEL files) | Number of normal samples | Number of disease samples | Platform information |
| GSE16515 | 16 | 36 | Affymetrix HG-U133 Plus 2.0, GPL570 |
| GSE15471 | 26 | 26 | Affymetrix HG-U133 Plus 2.0, GPL570 |
The rma function in the affy package of R language[12] was used to preprocess the raw data of the gene expression profiles, and the robust multi-array average (RMA) algorithm[13] was employed to calculate the amount of gene expression from the raw data of expression profiles and to obtain the gene expression values of the probes, that is, the signal strength values.
Using the Annotate package of R language combined with the chip annotation document, the probe sets were labeled with the corresponding genes (Entrez ID). In the situation of a single gene corresponding to multiple probe sets, the average value of the multiple probe sets was used to represent the expression value of the gene. In the case of multiple genes corresponding to one probe set, the probe set data were deleted.
The significance analysis of microarrays (SAM) algorithm[14] was used to screen for differentially expressed genes related to pancreatic cancer. The analysis platform employed the R language platform.
Gene Ontology (GO) analysis of differentially expressed genes was used to search for gene functions whose changes might be correlated with the differentially expressed genes of different samples[15,16]. The pathway analysis of differentially expressed genes was used to search for cellular pathways whose changes might be related to the differentially expressed genes in different samples[17-19]. The significance thresholds of enrichment analysis were 0.01 and 0.05 for GO analysis and pathway analysis, respectively. Descriptions of the result parameters of the functional annotations are shown in Supplementary Table 1.
Data screening and preprocessing:
Human protein interaction network data were downloaded from the STRING database[20], and protein interaction pairs with interaction scores of more than 900 were selected. Only protein interaction data containing differentially expressed genes were screened from the above qualified protein interaction data. Modules were mined and differentially expressed genes were annotated using the ClusterOne plugin of Cytoscape software.
Analysis of information crosstalk between interacting modules: The calculation method of crosstalk significance is as follows: in the context of a random network, the number of cases in which the number of interaction pairs between modules in N random networks (in this study, N = 1000) was greater than that in real networks was calculated and recorded as n. The formula for calculating the P value is P = n/N; a P value less than or equal to 0.05 represents significant crosstalk between modules.
Pivot analysis: The definition of pivot requires satisfaction of the following two conditions: (1) the pivot interacts with two modules at the same time and has at least two interaction pairs with each module; and (2) the P value of the significance analysis of the interaction between the pivot and each module should be less than or equal to 0.05. According to the above descriptions, the Python program was written to find the pivots between the functional modules, and the hypergeometric test method was used for the significance analysis.
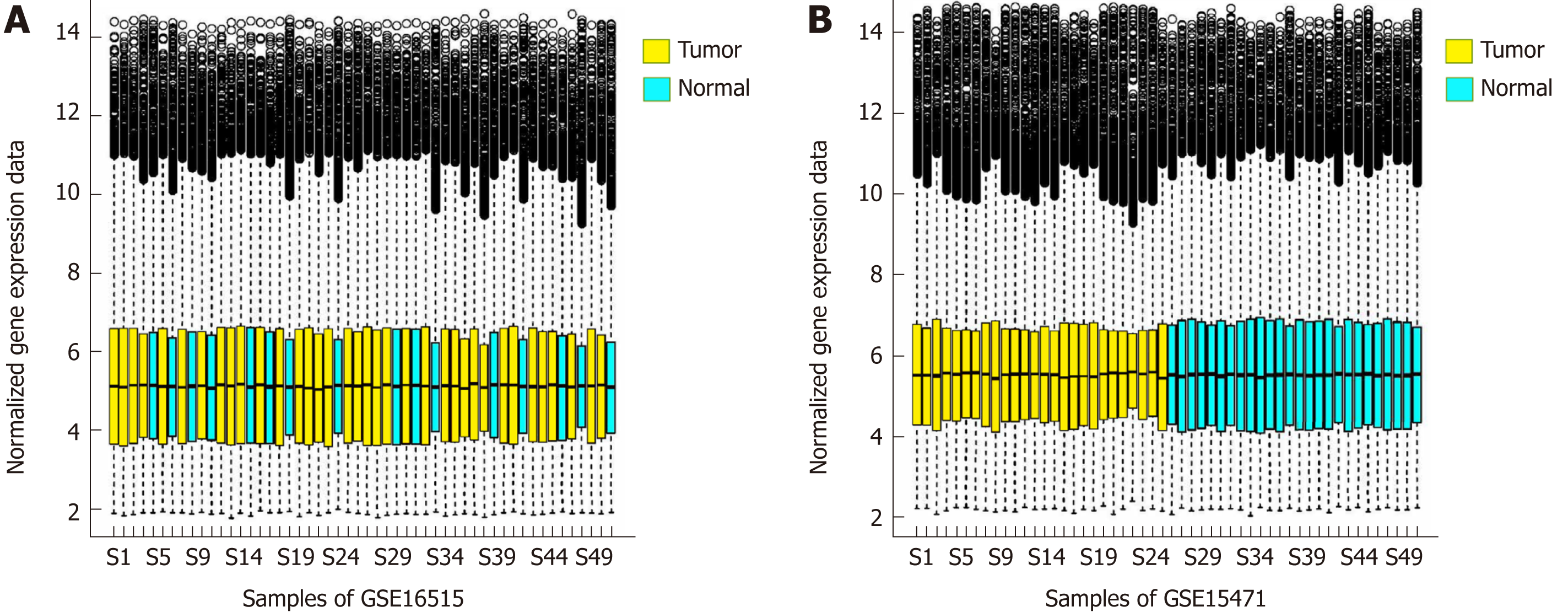
The distribution of gene expression amount calculated by the RMA algorithm is shown in Figure 1A and B After data preprocessing, the gene expression profile data were reduced from the original 54675 probe expression values to 20502 gene expression values.
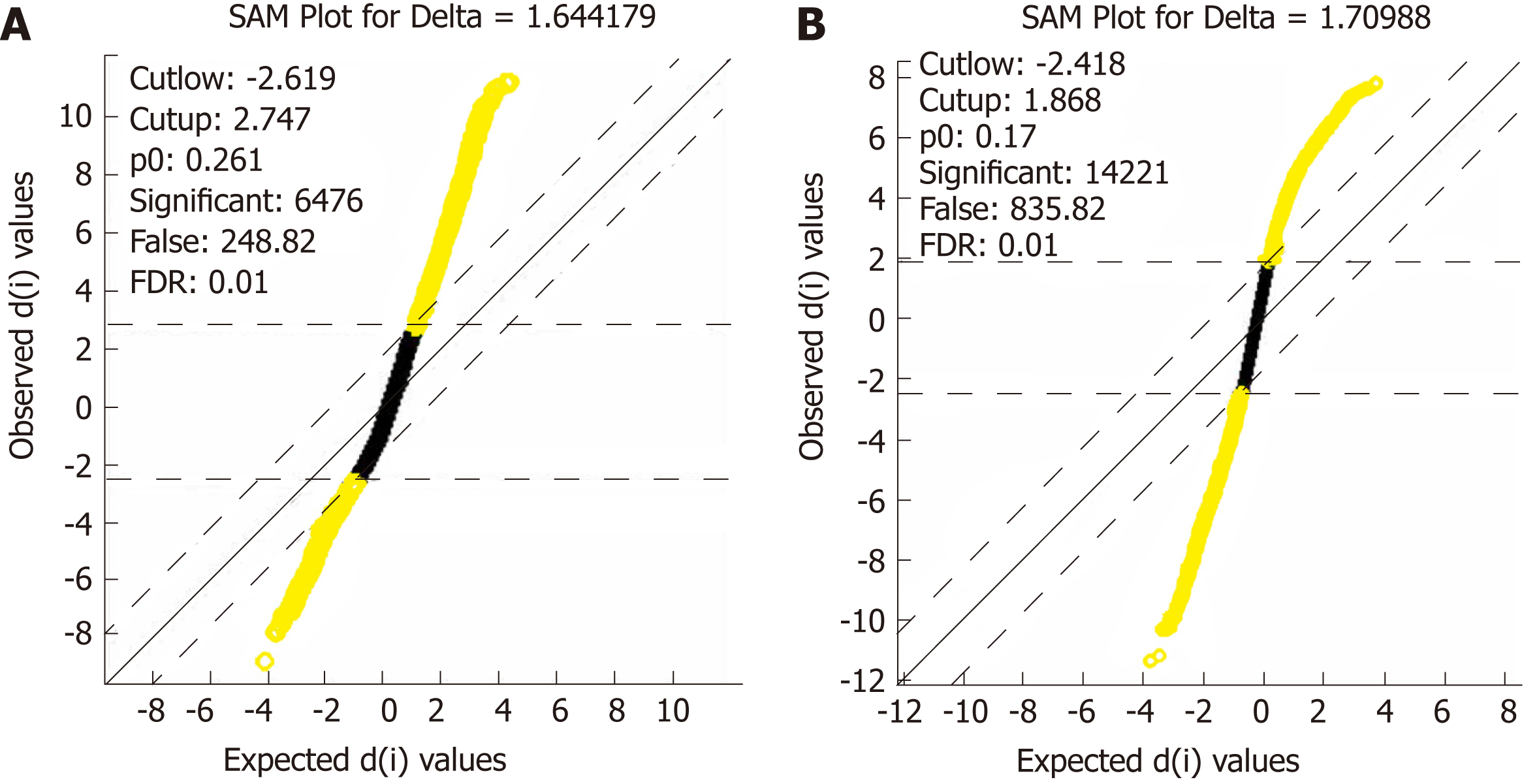
After data standardization and gene annotation, gene microarray significance analyses were performed on the two sets of data (GSE16515 and GSE15471) separately using the Sam function of the siggenes package of R language (Figure 2A and B); a total of 6098 and 12928 differentially expressed genes were obtained, respectively, and the first 40 genes were selected for display in Supplementary Tables 1 and 2, respectively. A total of 4870 core differentially expressed genes were obtained from the intersection of the two sets of differentially expressed genes for subsequent functional annotation analysis.
| Category | ID | Description |
| Biological processes | GO:2001236 | Regulation of the extrinsic apoptotic signaling pathway |
| GO:0097192 | Extrinsic apoptotic signaling pathway in the absence of ligand | |
| GO:0097191 | Extrinsic apoptotic signaling pathway | |
| GO:2001237 | Negative regulation of the extrinsic apoptotic signaling pathway | |
| GO:0031397 | Negative regulation of protein ubiquitination | |
| GO:0031396 | Regulation of protein ubiquitination | |
| GO:0010038 | Response to metal ions | |
| GO:0045862 | Positive regulation of proteolysis | |
| Cell composition | GO:0022624 | Proteasome accessory complex |
| GO:0034702 | Ion channel complex | |
| GO:0000502 | Proteasome complex | |
| GO:0005838 | Proteasome regulatory particle | |
| Molecular functions | GO:0003779 | Actin binding |
| GO:0002020 | Protease binding |
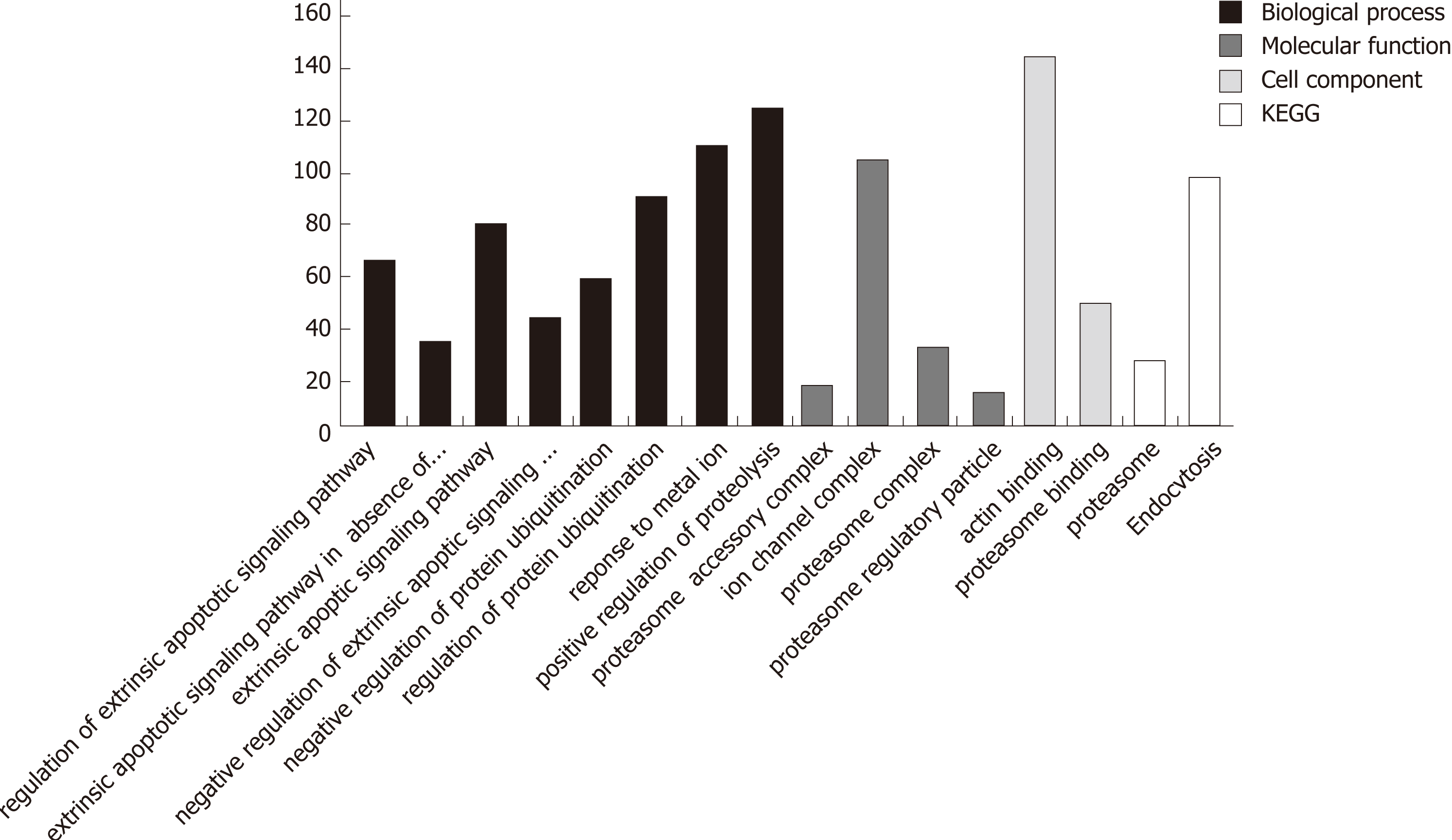
In the process of GO analysis of differentially expressed genes, the involvement of genes in biological processes, molecular functions and cell compositions was annotated by setting different parameters. The 14 functional items related to apoptosis/autophagy are listed in Table 2.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis enriched the differentially expressed genes into four pathways: hsa03050, hsa04144, hsa05412 and hsa04020. hsa04144 is related to endocytosis, hsa04020 is related to the calcium signaling pathway, and both are, to a certain degree, related to autophagy.
Searching the GENE functional annotation database of the NCBI using three keywords, autophagy, Homo sapiens and cancer (adopt and logic rule), 851 genes were obtained. The 16 functional items associated with autophagy obtained by GO and KEGG analyses revealed a total of 986 differentially expressed genes (Figure 3). After removing genes that are clearly defined in the GENE database, 347 differentially expressed genes were obtained (Supplementary Table 3). The relationship between these genes and the autophagy of pancreatic cancer cells awaits further exploration.
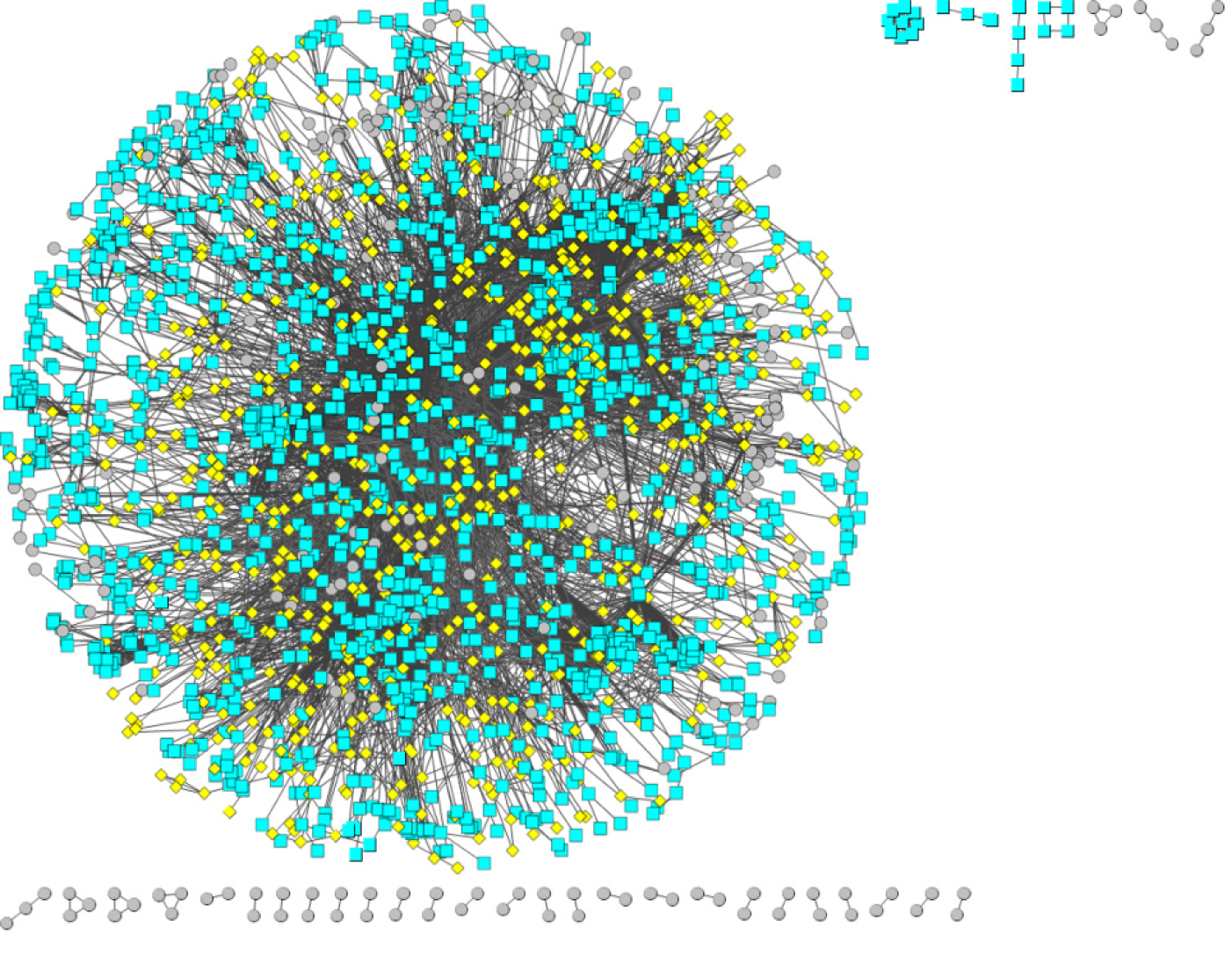
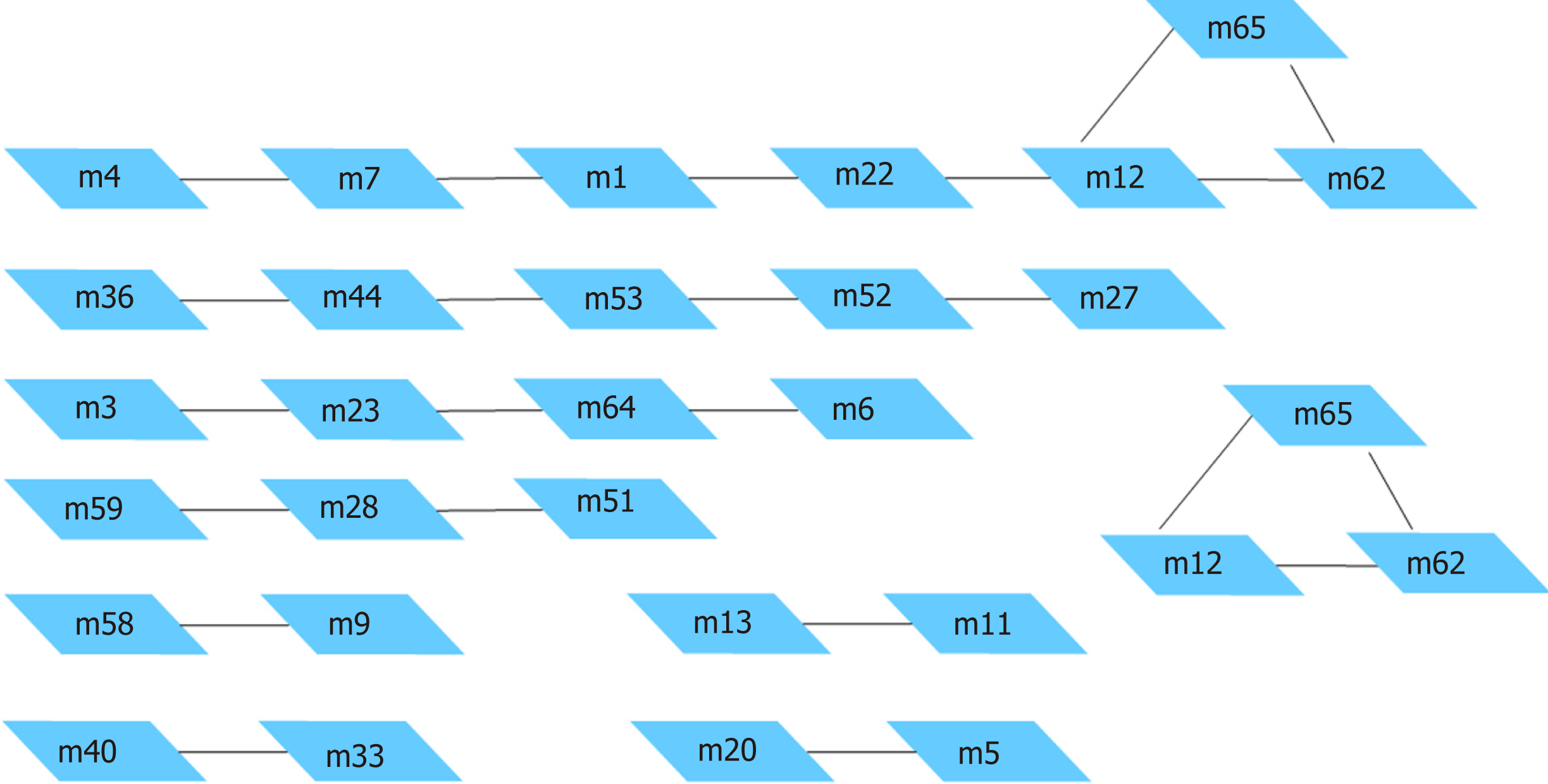
Based on a protein interaction network database (STRING), a reliable protein interaction network of 10524 proteins and 196254 interaction pairs was obtained by selecting protein interaction pairs with an interaction score of greater than or equal to 900. Protein interaction data containing only differentially expressed genes were subsequently screened from the above protein interaction data, and a protein interaction network of 2256 differentially expressed genes and 12632 interaction pairs was obtained. Figure 4 shows the protein interaction network composed of differentially expressed genes. Red pivot dots indicate that when mining modules, the P values of the participating modules were less than 0.05, while yellow pivot dots indicate that when mining modules, the P values of the participating modules were greater than 0.05. Grey pivot dots indicate that these genes were not involved in the modules. A total of 65 clustering modules were screened by importing the above interaction data containing differentially expressed genes into Cytoscape (Supplementary Table 4).
GO and KEGG functional annotation of the differentially expressed genes in the protein interaction network was performed. The results are shown in Supplementary Tables 5 and 6. GO analysis found that GO:0016236 was involved in autophagy, while KEGG analysis revealed a highly significant (P = 1.06E-07) new pathway, hsa04216 (ferroptosis), a new cell death pattern associated with iron death.
Next, we identified the MAP1L3A (LC3) gene in the excavated module 32, which is the target gene of emphasis in this study. We also found that this module contains multiple genes related to autophagy (Supplementary Table 7). Through the functional enrichment analysis of module 32, the genes of this module were found to be mainly involved in pathways related to autophagy and iron-dependent cell death.
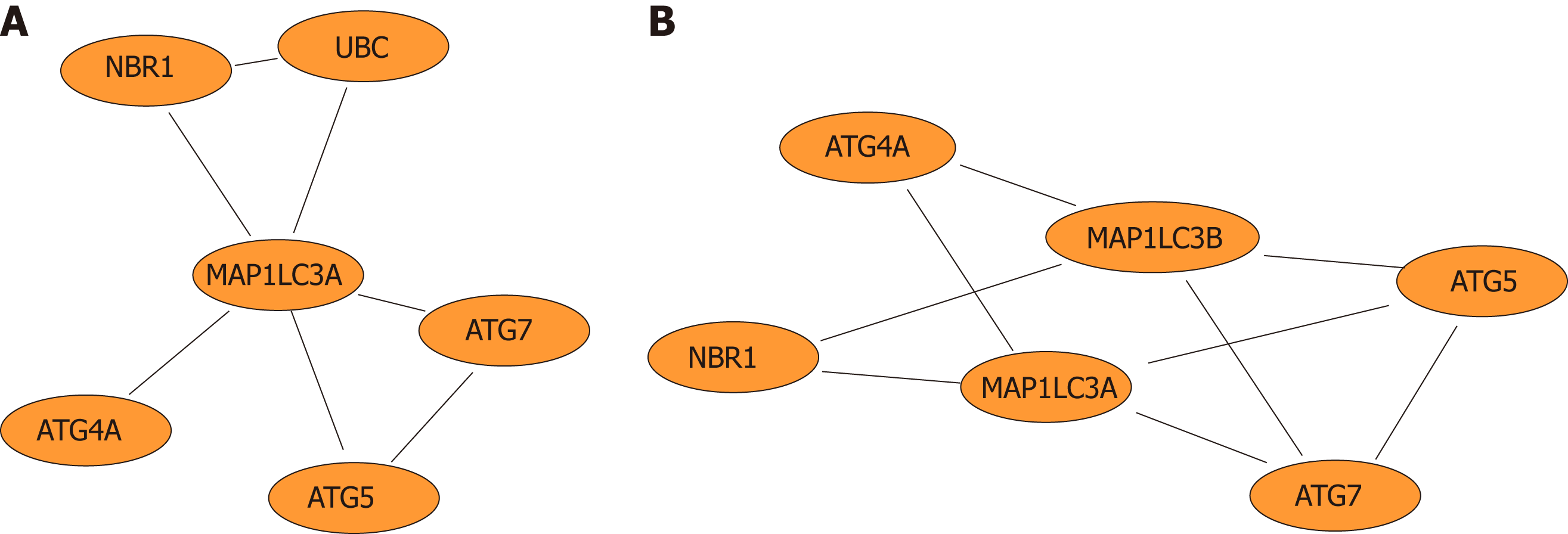
Furthermore, network diagrams were used to show the LC3 gene and genes that directly interact with LC3 (Figure 5A) and to demonstrate the functional modules in which the LC3 gene is involved (Figure 5B).
The complex intracellular pathway networks were analyzed by crosstalk, and important protein pivots that affect signal transduction between the pathways were identified through protein interactions. The results of the pathway crosstalk analysis of the aforementioned clustering modules are shown in Supplementary Table 8 and Figure 6.
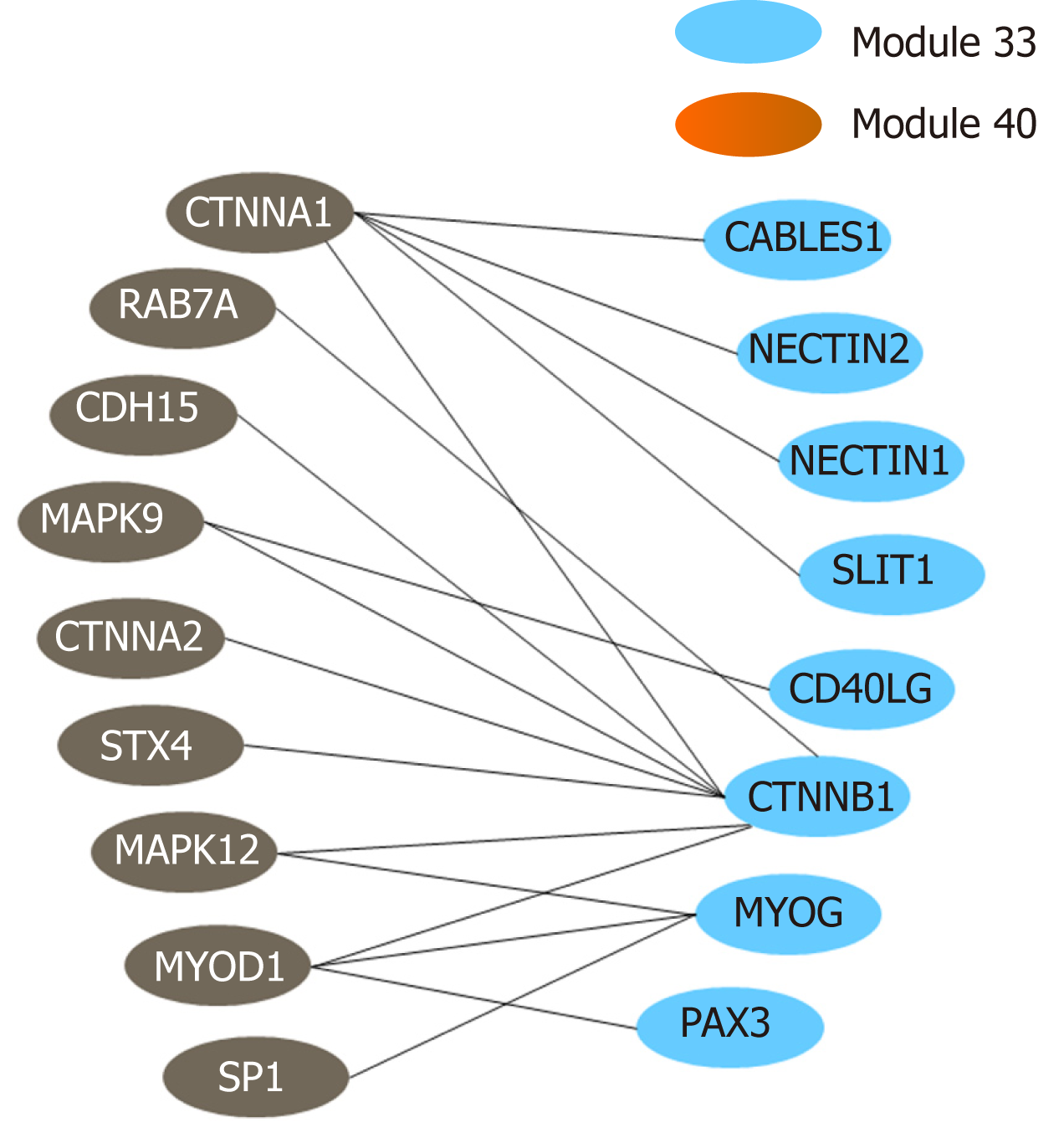
The above functional annotation analysis revealed that modules 33 and 40 were both associated with autophagy; the crosstalk between the two modules is demonstrated using a network diagram by Cytoscape (Figure 7). The genes involved in the two modules and their functional descriptions are listed in Supplementary Table 9.
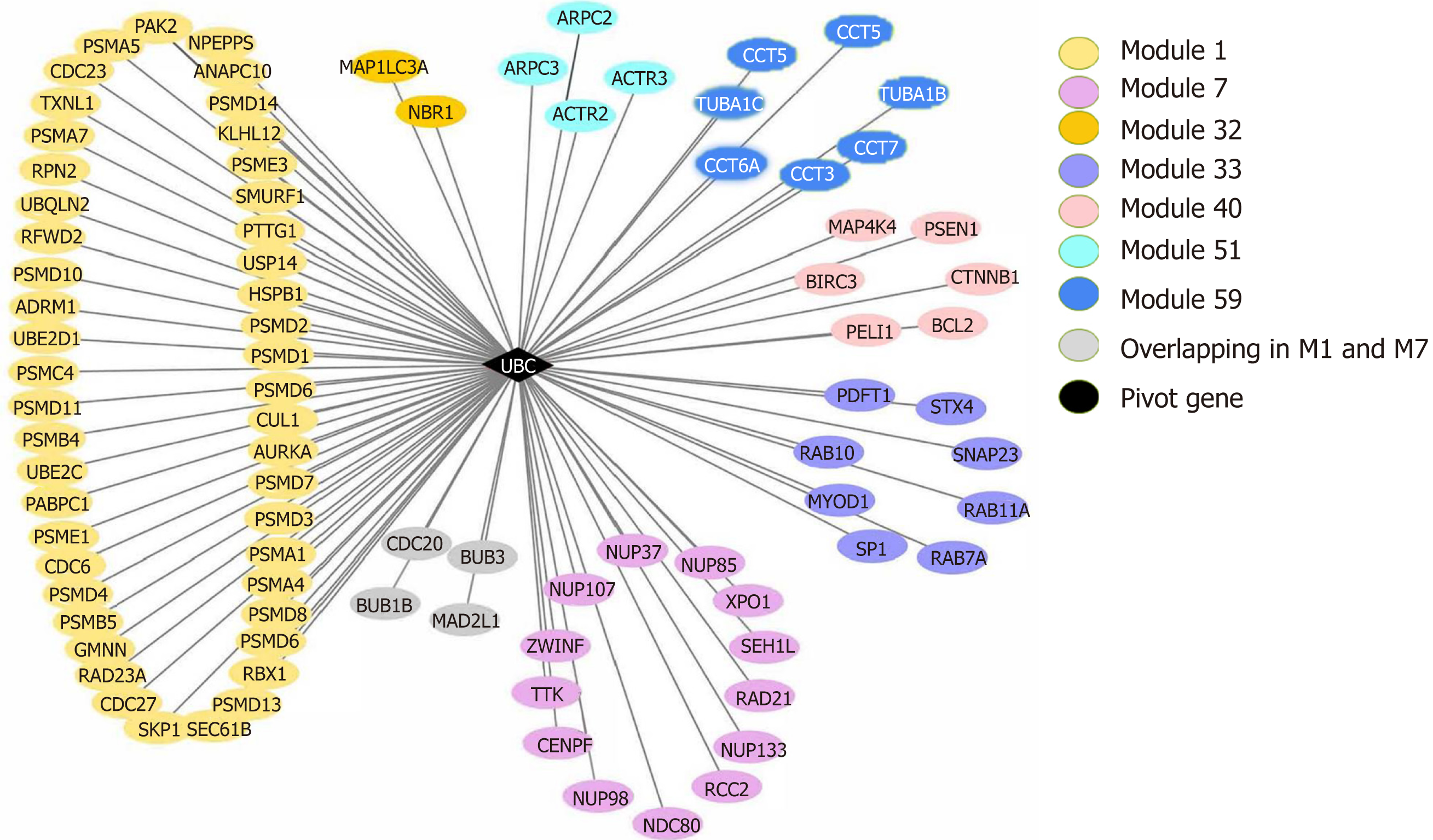
Pivot analysis revealed that the gene 7316, ubiquitin C (UBC), was associated with several modules related to cancer and autophagy. After removing the redundant modules, seven modules interacting with this pivot were obtained (Supplementary Table 10). These modules all contain genes that directly interact with gene 7316. Functional enrichment analysis showed that most of these 7 modules are related to human cell death, especially the autophagy process. In module 32, the pivot directly interacts with gene 84557 (MAP1LC3A) and is associated with multiple modules (Figure 8).
Pancreatic cancer has a high malignant degree, a low early diagnosis rate, and a less than 20% chance of radical resection. The incidence and mortality rate of pancreatic cancer are high. From 2000 to 2014, the overall 5-year survival rate of pancreatic cancer in different regions of the world was between 5% and 15%[21]. In China, approximately 90100 new cases of pancreatic cancer were reported in 2015, ranking ninth among all malignant tumors, while mortality reached 79400 cases, ranking sixth among all malignant tumors; the 5-year survival rate was 9.9%[22]. Until now, an effective diagnosis and treatment for pancreatic cancer have been lacking, and the exact causes and pathogenesis of pancreatic cancer are not well understood. In this study, bioinformatics analysis based on the LC3 gene was used to identify differentially expressed genes in pancreatic cancer and the results were comprehensively analyzed to provide a new experimental basis for the diagnosis and treatment of pancreatic cancer.
First, the GO analysis results of the differentially expressed genes associated with pancreatic cancer were preliminarily studied, and 347 genes that had not been clearly defined by the GENE database as directly related to the autophagy of human cancer cells were identified, providing new information and a novel direction for future cancer and autophagy research. Previous studies have shown that the occurrence and development of pancreatic cancer are the result of the interaction and influence of multiple genes and factors[23,24]. In this study, GO functional annotation analysis revealed that functional annotations were associated with multiple differentially expressed genes, indirectly confirming the earlier results. This study was not limited to an expression abnormality or a mutation of a single gene or protein but rather analyzed the gene microarray expression profile data to obtain the suspected differentially expressed genes associated with tumors, providing useful data for future studies and providing a reliable and detailed experimental basis for the diagnosis and treatment of complex diseases. Second, the KEGG analysis results of pancreatic cancer-related differentially expressed genes were analyzed. The differentially expressed genes were enriched in four pathways, and the complex molecular mechanisms involved in these pathways are related to the occurrence of tumors, inflammation, immune disorders, and idiopathic diseases[25].
In this study, we combined the gene expression database and the STRING protein interaction database to mine modules in the protein interaction network involving the differentially expressed genes and found that modules 33 and 40 were both associated with autophagy. In view of the functional descriptions, the genes involved in these two modules are closely related to cellular processes such as cancer, autophagy and apoptosis. Through functional enrichment analysis of the genes involved in these modules, KEGG enrichment further identified pathway hsa04216, that is, ferroptosis[26], which is characterized by the production of reactive oxygen species (ROS) from the peroxidation of accumulated iron and lipids. Kang et al[27] reviewed the relationship between autophagy and ferroptosis and noted that the activation of ferroptosis depends on its induction by autophagy.
In the module interaction study, we found that UBC, as a pivot of module interactions, connected several modules related to cancer and autophagy and plays an important role in multiple cell death modules related to autophagy (Figures 1-8), consistent with current knowledge and our understanding of cancer and cell death. UBC, as a precursor of ubiquitin, plays a key role in the occurrence and development of diseases such as autophagy, cancer, and inflammation through the ubiquitin proteasome system (UPS)[28]. A large number of studies have shown a close relationship between UPS and autophagy[29,30]. Previously, UPS and autophagy have been considered complementary degradation systems with no intersection. However, some monoubiquitinated proteins can also be degraded by autophagy[31]. Pandey et al[32] performed in vitro experiments and showed that monoubiquitination and histone deacetylase 6 (HDAC6) are the key signals linking the two systems of autophagy and UPS, consistent with the results of the current study that the ubiquitin precursor UBC connects multiple autophagy-related modules.
A previous study showed that the signaling pathway adapter protein P62 contains an LC3 recognition sequence that forms oligomers to recruit ubiquitinated proteins through a ubiquitin binding domain and interacts with LC3 to form degradation substrates for the autophagosome[33]. Ubiquitin is also one of the substrates of molecular chaperone-mediated autophagy[34]. In this study, a direct interaction between LC3 and the pivot gene UBC was identified, which has certain significance for the study of the mechanism by which autophagy scavenges ubiquitinated substrates. Ubiquitin and UPS are also closely related to the occurrence and development of tumors. Tang et al[35] confirmed that ubiquitin is highly expressed in many types of tumor tissues. Liu et al[36] showed that UPS could selectively degrade the products of oncogenes and tumor suppressor genes, as well as apoptosis-regulating proteins, thus regulating cell mutation and tumorigenesis.
In recent years, the perineural invasion (PNI) of pancreatic cancer has become a research hotspot in academia and in the clinic; however, its mechanism has not yet been elucidated. A complicated tumor microenvironment, autophagy, and neural plasticity[37] exert important effects on the nerves around and inside the pancreas. It has been reported[38] that pancreatic cancer tissue with high expression of ubiquitin-specific protease 9X (USP9X), a member of the ubiquitin-specific protease subfamily of the family of deubiquitinating enzymes, may be more invasive, and high expression of USP9X is closely associated with the prognosis of pancreatic cancer. In UPS, the reversible process of protein ubiquitination and deubiquitination catalyzed by ubiquitinating enzymes (UBEs) and deubiquitinating enzymes (DUBs) is also closely related to PNI of the tumor[39]. UBC-terminal hydrolases (UCHs) are a subfamily of DUBs. Previous studies have shown that the UCH family plays different roles in the progression of different tumors[40]. Ubiquitin carboxyl terminal esterase L1, a family member of UCHs, is not only overexpressed in neural tissue[41] but is also used as a marker of nerve fibers to study PNI[42].
In the study of modular clustering based on protein interactions, UBC was clustered with multiple autophagy-related genes in module 32 and directly interacted with LC3 and NBR1 (Figures 2-7). UCH uses ubiquitinated proteins as substrates to catalyze the removal of ubiquitin molecules. Therefore, UBC and UCH also directly interact. Moreover, the expression level of UCH is closely related to the process of PNI. Based on our previous study[6] in which the positive rate of PNI was significantly positively correlated with the high expression of LC3 in pancreatic cancer patients and that PNI and LC3 levels are independent risk factors for the poor prognosis of pancreatic cancer, we postulate that the autophagy-related protein LC3 may establish a close association with pancreatic cancer PNI through UBC and its associated ubiquitin proteasome system.
In summary, the present study combined the gene expression profile microarray technique with bioinformatics analysis technology to analyze and mine a large quantity of data and identified new significantly differentially expressed genes related to the occurrence of autophagy in pancreatic cancer, which expands the autophagy-related gene profile of pancreatic cancer and is helpful for the search of candidate susceptible genes and rare mutations that may be associated with the occurrence and development of autophagy in pancreatic cancer. The identification and review of UBC, a key gene that directly interacts with LC3, suggest that it may be a key factor that leads to a poor prognosis of pancreatic cancer mediated by PNI and suggests a new direction for further research.
In this study, we identified differentially expressed genes between pancreatic cancer cells and normal cells at the whole-genome level using the whole-genome expression profiling technique, identified 347 genes that have no confirmed association with the autophagy process of human pancreatic cancer cells in previous studies, and discovered and clarified information about the pathways involved in autophagy. Furthermore, we identified UBC, which plays an important role in several modules related to cell death, through gene expression microarray analysis based on autophagy and LC3 and found that UBC is widely involved in tumor cell growth, invasion and metastasis. It was also found that ubiquitin is closely related to PNI. It is believed that LC3 may affect the PNI and prognosis of pancreatic cancer through UBC, which is helpful for the study and treatment of pancreatic cancer and provides an important clue to the pathogenesis of pancreatic cancer.
Pancreatic cancer is a malignant tumor with a poor prognosis that has almost equal mortality and morbidity in patients. At present, more and more studies have found that autophagy is closely related to the occurrence, development, differentiation and prognosis of pancreatic cancer. Autophagy-related protein microtubule-associated protein 1A/1B-light chain 3 (LC3), as a key protein in the autophagy process, is involved in the formation of the autophagosome. A study by our research group found that high expression of LC3 in pancreatic cancer was positively correlated with neural invasion and poor prognosis. With the development of genomics, gene expression microarray technology, proteomics and bioinformatics, the ability to manipulate and characterize human genes and their products has been acquired. Disease-related genes have been studied at the molecular level to understand the pathogenesis of diseases. On the basis of previous studies and using LC3 as a guidance index, the autophagy gene expression profile of pancreatic cancer was analyzed to guide the functional annotation of differentially expressed genes and to enhance the reliability of bioinformatics prediction and analysis. Thus, to provide a basis for the study of the molecular mechanism of autophagy in pancreatic cancer.
This study focused on the differentially expressed genes based on LC3 to analyze the gene expression profile of autophagy in pancreatic cancer. Thus, to provide a basis for the study of the molecular mechanism of autophagy in pancreatic cancer.
To identify differentially expressed genes in autophagy of pancreatic cancer and to provide a basis for exploring the molecular mechanism of autophagy of pancreatic cancer cells and finding new targets for diagnosis and treatment of pancreatic cancer.
On the basis of previous studies and using LC3 as a guidance index, differentially expressed genes involved in the autophagy of pancreatic cancer were identified by a gene expression microarray technique. Protein interaction networks were constructed and the functional clustering of differentially expressed genes was carried out. Key interacting proteins or genes between modules were screened and evaluated by statistical methods, and the pathogenesis of pancreatic cancer was explored.
After removing genes that are clearly defined in the GENE database, 347 differentially expressed genes were obtained. The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed a highly significant new pathway, hsa04216 (ferroptosis), a new cell death pattern associated with iron death. The ubiquitin C (UBC) as a pivot of module interactions, connected several modules related to cancer and autophagy and plays an important role in multiple cell death modules related to autophagy.
In this study, we identified differentially expressed genes based on the LC3 to analyze the gene expression profile of autophagy in pancreatic cancer. Three hundred and forty-seven genes that have no confirmed association with the autophagy process of human pancreatic cancer cells in previous studies were concentrated, and the key pathways involved in autophagy were enriched. Furthermore, a key gene UBC which is closely related to the occurrence of perineural invasion (PNI) was determined, suggesting that LC3 may influence the PNI and prognosis of pancreatic cancer through UBC.
With the development of genomics, gene expression microarray technology, proteomics and bioinformatics, we have been able to study disease-related genes at the molecular level to understand the pathogenesis of disease, thus to seek new research directions or to find new targets for clinical diagnosis and treatment. In this study, LC3 was used as a target to explore the differential genes related to autophagy in pancreatic cancer cells. Three hundred and forty-seven genes that have no confirmed association with the autophagy process of human pancreatic cancer cells in previous studies were concentrated, it is obviously unrealistic to analyze all the genes interacting with LC3 in vitro. Nevertheless, a key gene UBC which is closely related to the occurrence of PNI was determined. Our previous results showed that the high expression of LC3 was positively correlated with PNI in the patients with pancreatic cancer. Suggesting that LC3 may influence the PNI and prognosis of pancreatic cancer through UBC. Therefore, we have planned to supplement some vitro and vivo experiments to further analysis the relationship between them and explore the molecular mechanism of phagocytosis in pancreatic cancer cells.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Muhammad JS, Zhang ZY S-Editor: Ma RY L-Editor: A E-Editor: Ma YJ
| 1. | Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73-85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1397] [Cited by in F6Publishing: 1562] [Article Influence: 195.3] [Reference Citation Analysis (0)] |
| 2. | Brech A, Ahlquist T, Lothe RA, Stenmark H. Autophagy in tumour suppression and promotion. Mol Oncol. 2009;3:366-375. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 133] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 3. | Zhou Y, Zhou Q, Chen R. Pancreatic stellate cells promotes the perineural invasion in pancreatic cancer. Med Hypotheses. 2012;78:811-813. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Tuloup-Minguez V, Greffard A, Codogno P, Botti J. Regulation of autophagy by extracellular matrix glycoproteins in HeLa cells. Autophagy. 2011;7:27-39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Guo K, Ma Q, Li J, Wang Z, Shan T, Li W, Xu Q, Xie K. Interaction of the sympathetic nerve with pancreatic cancer cells promotes perineural invasion through the activation of STAT3 signaling. Mol Cancer Ther. 2013;12:264-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 6. | Yang YH, Liu JB, Gui Y, Lei LL, Zhang SJ. Relationship between autophagy and perineural invasion, clinicopathological features, and prognosis in pancreatic cancer. World J Gastroenterol. 2017;23:7232-7241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 27] [Cited by in F6Publishing: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Tsugata T, Nikoh N, Kin T, Saitoh I, Noguchi Y, Ueki H, Watanabe M, James Shapiro AM, Noguchi H. Potential Factors for the Differentiation of ESCs/iPSCs Into Insulin-Producing Cells. Cell Med. 2014;7:83-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 8. | Artemov A, Aliper A, Korzinkin M, Lezhnina K, Jellen L, Zhukov N, Roumiantsev S, Gaifullin N, Zhavoronkov A, Borisov N, Buzdin A. A method for predicting target drug efficiency in cancer based on the analysis of signaling pathway activation. Oncotarget. 2015;6:29347-29356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | Zhao LP, Li RH, Han DM, Zhang XQ, Nian GX, Wu MX, Feng Y, Zhang L, Sun ZG. Independent prognostic Factor of low-expressed LncRNA ZNF667-AS1 for cervical cancer and inhibitory function on the proliferation of cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21:5353-5360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 17] [Reference Citation Analysis (0)] |
| 10. | Fei HJ, Chen SC, Zhang JY, Li SY, Zhang LL, Chen YY, Chang CX, Xu CM. Identification of significant biomarkers and pathways associated with gastric carcinogenesis by whole genome-wide expression profiling analysis. Int J Oncol. 2018;52:955-966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Available from: http://www.ncbi.nlm.nih.gov/Web/Genbank/. [PubMed] [Cited in This Article: ] |
| 12. | Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3737] [Cited by in F6Publishing: 3869] [Article Influence: 193.5] [Reference Citation Analysis (0)] |
| 13. | Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6199] [Cited by in F6Publishing: 6048] [Article Influence: 288.0] [Reference Citation Analysis (0)] |
| 14. | Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116-5121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9125] [Cited by in F6Publishing: 9571] [Article Influence: 416.1] [Reference Citation Analysis (0)] |
| 15. | Amar D, Vizel A, Levy C, Shamir R. ADEPTUS: a discovery tool for disease prediction, enrichment and network analysis based on profiles from many diseases. Bioinformatics. 2018;34:1959-1961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Wang J, Du Q, Li C. Bioinformatics analysis of gene expression profiles to identify causal genes in luminal B2 breast cancer. Oncol Lett. 2017;14:7880-7888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Huo T, Canepa R, Sura A, Modave F, Gong Y. Colorectal cancer stages transcriptome analysis. PLoS One. 2017;12:e0188697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Tello-Ruiz MK, Naithani S, Stein JC, Gupta P, Campbell M, Olson A, Wei S, Preece J, Geniza MJ, Jiao Y, Lee YK, Wang B, Mulvaney J, Chougule K, Elser J, Al-Bader N, Kumari S, Thomason J, Kumar V, Bolser DM, Naamati G, Tapanari E, Fonseca N, Huerta L, Iqbal H, Keays M, Munoz-Pomer Fuentes A, Tang A, Fabregat A, D'Eustachio P, Weiser J, Stein LD, Petryszak R, Papatheodorou I, Kersey PJ, Lockhart P, Taylor C, Jaiswal P, Ware D. Gramene 2018: unifying comparative genomics and pathway resources for plant research. Nucleic Acids Res. 2018;46:D1181-D1189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 19. | Zhao R, Wang Y, Zhang M, Gu X, Wang W, Tan J, Wei X, Jin N. Screening of potential therapy targets for prostate cancer using integrated analysis of two gene expression profiles. Oncol Lett. 2017;14:5361-5369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808-D815. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3099] [Cited by in F6Publishing: 3198] [Article Influence: 290.7] [Reference Citation Analysis (0)] |
| 21. | Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, Ogunbiyi OJ, Azevedo E Silva G, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP; CONCORD Working Group. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023-1075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2711] [Cited by in F6Publishing: 2730] [Article Influence: 455.0] [Reference Citation Analysis (0)] |
| 22. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11444] [Cited by in F6Publishing: 12540] [Article Influence: 1567.5] [Reference Citation Analysis (0)] |
| 23. | Furukawa T, Horii A. Molecular pathology of pancreatic cancer: in quest of tumor suppressor genes. Pancreas. 2004;28:253-256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Slebos RJ, Hoppin JA, Tolbert PE, Holly EA, Brock JW, Zhang RH, Bracci PM, Foley J, Stockton P, McGregor LM, Flake GP, Taylor JA. K-ras and p53 in pancreatic cancer: association with medical history, histopathology, and environmental exposures in a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1223-1232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 25. | Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 360] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 26. | Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-1072. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4711] [Cited by in F6Publishing: 8258] [Article Influence: 688.2] [Reference Citation Analysis (1)] |
| 27. | Kang R, Tang D. Autophagy and Ferroptosis - What's the Connection? Curr Pathobiol Rep. 2017;5:153-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 28. | Shi D, Grossman SR. Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol Ther. 2010;10:737-747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 29. | Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363-397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1008] [Cited by in F6Publishing: 1074] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 30. | Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, Harris AL. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415-4423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 227] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 31. | Kaminskyy V, Zhivotovsky B. Proteases in autophagy. Biochim Biophys Acta. 2012;1824:44-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 32. | Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859-863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 862] [Cited by in F6Publishing: 916] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 33. | Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843:163-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 34. | Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727-738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1102] [Cited by in F6Publishing: 1120] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 35. | Tang Y, Geng Y, Luo J, Shen W, Zhu W, Meng C, Li M, Zhou X, Zhang S, Cao J. Downregulation of ubiquitin inhibits the proliferation and radioresistance of non-small cell lung cancer cells in vitro and in vivo. Sci Rep. 2015;5:9476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 36. | Liu J, Shaik S, Dai X, Wu Q, Zhou X, Wang Z, Wei W. Targeting the ubiquitin pathway for cancer treatment. Biochim Biophys Acta. 2015;1855:50-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 88] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 37. | Demir IE, Friess H, Ceyhan GO. Neural plasticity in pancreatitis and pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2015;12:649-659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 38. | Liu L, Yao D, Zhang P, Ding W, Zhang X, Zhang C, Gong S, Zhang Y, Wang J, Sun T, Ren Z. Deubiquitinase USP9X promotes cell migration, invasion and inhibits apoptosis of human pancreatic cancer. Oncol Rep. 2017;38:3531-3537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 39. | Neutzner M, Neutzner A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012;52:37-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 40. | Fang Y, Shen X. Ubiquitin carboxyl-terminal hydrolases: involvement in cancer progression and clinical implications. Cancer Metastasis Rev. 2017;36:669-682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 41. | Wang Y, Wang L, Gao H, Gao Y, Yang C, Ji H, Dong W. UCHL1 expression and localization on testicular development and spermatogenesis of Chinese giant salamanders. Oncotarget. 2017;8:86043-86055. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 42. | Li J, Ma Q, Liu H, Guo K, Li F, Li W, Han L, Wang F, Wu E. Relationship between neural alteration and perineural invasion in pancreatic cancer patients with hyperglycemia. PLoS One. 2011;6:e17385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |